circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000180376:+:3:56566593:56567053
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000180376:+:3:56566593:56567053 | ENSG00000180376 | MSTRG.23130.17 | + | 3 | 56566594 | 56567053 | 270 | GUCAAUAUAGUCUAUAUUUAAACAGUAUUUCUAAUCAGCCAAAGGAUGAGAACAUUAUGGGAUUAUUCAAAAAAACUGAAAUGGUUUCAUCUGUCCCAGCUGAAAAUAAAUCUGUCUUAAAUGAACAUCAGGAGACAUCUAAACAGUGUGAGCAAAAAAUUGCCAUAGAGAAUGAAUGGAAACCAGCUGAUAUAUUCAGUACUCUGGGGGAAAGGGAAUGUGAUAGAAGUUCGUUGGAAGCAAAAAAAGCCCAGUGGAGGAAAGAGCUAGGUCAAUAUAGUCUAUAUUUAAACAGUAUUUCUAAUCAGCCAAAGGAUGAG | circ |
| ENSG00000180376:+:3:56566593:56567053 | ENSG00000180376 | MSTRG.23130.17 | + | 3 | 56566594 | 56567053 | 22 | GAAAGAGCUAGGUCAAUAUAGU | bsj |
| ENSG00000180376:+:3:56566593:56567053 | ENSG00000180376 | MSTRG.23130.17 | + | 3 | 56566394 | 56566603 | 210 | GGACUACAGGUGUGAGCCACCACUCCCUGCUUCCUUUUUUUCUUUUUGACAGGAAGGGAAACUUAAUGGCUACCUCAAGGAUUGUUUUGAGAGUUAAUUUAAGUAACGUGUAAGACAUGUAAGAACUGUAUAGCACUAUGCCAAGUAUUAUAACAGAUGUAAUUUAGUGUUAAUUUUAAAACAUGUAUUUUACCUCCUAGGUCAAUAUAG | ie_up |
| ENSG00000180376:+:3:56566593:56567053 | ENSG00000180376 | MSTRG.23130.17 | + | 3 | 56567044 | 56567253 | 210 | AAAGAGCUAGGUAGGUAACUUUUAUACCUUUAUUAUAGUUUUAUUGGCUUAAAGAAUAUGUAUUGAAGCCGGGCACGGUGGCUCACACCUGUAAUACCAGCACUUUGGGAAGCCGAGGUGGGUGGAUCAGGAGGGCAAGAGAUCAAGACCAUCCUGGCCAACAUGGUGAAAUCCCAUCUCUACUAAAAAUAUAAAAAUUAGCUGGGCGUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
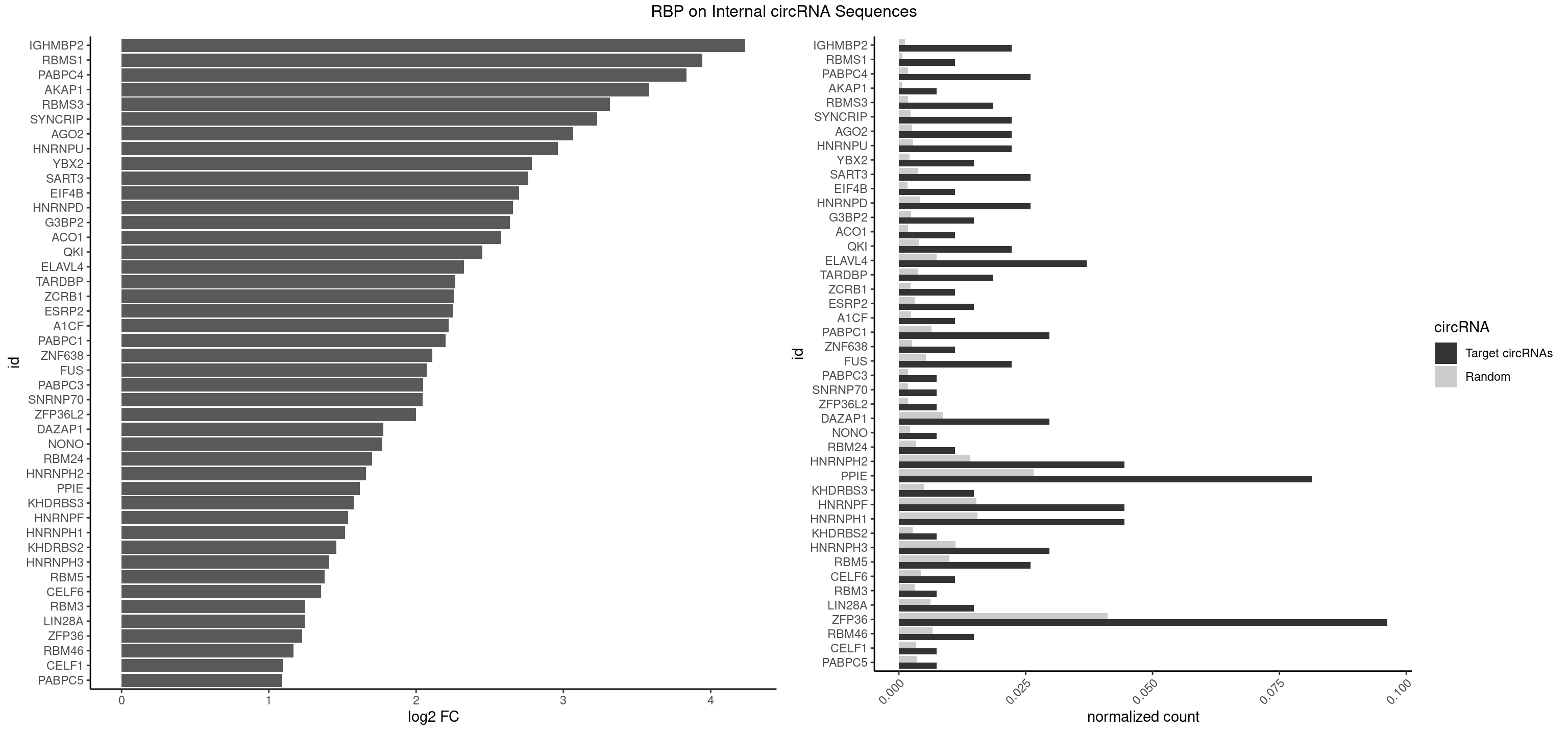
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 5 | 813 | 0.022222222 | 0.0011796520 | 4.235570 | AAAAAA | AAAAAA |
| RBMS1 | 2 | 497 | 0.011111111 | 0.0007217036 | 3.944453 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PABPC4 | 6 | 1251 | 0.025925926 | 0.0018144033 | 3.836828 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| AKAP1 | 1 | 426 | 0.007407407 | 0.0006188101 | 3.581400 | AUAUAU | AUAUAU,UAUAUA |
| RBMS3 | 4 | 1283 | 0.018518519 | 0.0018607779 | 3.314991 | AAUAUA,AUAUAG,AUAUAU,CUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SYNCRIP | 5 | 1634 | 0.022222222 | 0.0023694485 | 3.229380 | AAAAAA | AAAAAA,UUUUUU |
| AGO2 | 5 | 1830 | 0.022222222 | 0.0026534924 | 3.066039 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPU | 5 | 1965 | 0.022222222 | 0.0028491350 | 2.963407 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| YBX2 | 3 | 1480 | 0.014814815 | 0.0021462711 | 2.787136 | AACAUC,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SART3 | 6 | 2634 | 0.025925926 | 0.0038186524 | 2.763260 | AAAAAA,AAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| EIF4B | 2 | 1179 | 0.011111111 | 0.0017100607 | 2.699884 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPD | 6 | 2837 | 0.025925926 | 0.0041128408 | 2.656188 | AAAAAA,UAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| G3BP2 | 3 | 1644 | 0.014814815 | 0.0023839405 | 2.635620 | AGGAUG,GGAUGA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ACO1 | 2 | 1283 | 0.011111111 | 0.0018607779 | 2.578025 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| QKI | 5 | 2805 | 0.022222222 | 0.0040664663 | 2.450156 | AUCUAA,CUAAUC,UAAUCA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ELAVL4 | 9 | 5105 | 0.037037037 | 0.0073996354 | 2.323443 | AAAAAA,AUCUAA,UAUUUA,UCUAAU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TARDBP | 4 | 2654 | 0.018518519 | 0.0038476365 | 2.266924 | GAAUGA,GAAUGG,GAAUGU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZCRB1 | 2 | 1605 | 0.011111111 | 0.0023274215 | 2.255199 | AUUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| ESRP2 | 3 | 2150 | 0.014814815 | 0.0031172377 | 2.248701 | GGGAAA,GGGAAU,GGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| A1CF | 2 | 1642 | 0.011111111 | 0.0023810421 | 2.222338 | CAGUAU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC1 | 7 | 4443 | 0.029629630 | 0.0064402624 | 2.201849 | AAAAAA,AAAAAC,CUAAUC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ZNF638 | 2 | 1773 | 0.011111111 | 0.0025708878 | 2.111665 | CGUUGG,GUUCGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| FUS | 5 | 3648 | 0.022222222 | 0.0052881452 | 2.071169 | AAAAAA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| PABPC3 | 1 | 1234 | 0.007407407 | 0.0017897669 | 2.049197 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 1 | 1237 | 0.007407407 | 0.0017941145 | 2.045697 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZFP36L2 | 1 | 1277 | 0.007407407 | 0.0018520827 | 1.999820 | UAUUUA | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| DAZAP1 | 7 | 5964 | 0.029629630 | 0.0086445016 | 1.777186 | AAAAAA,AGGAAA,AGGAUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| NONO | 1 | 1498 | 0.007407407 | 0.0021723567 | 1.769708 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM24 | 2 | 2357 | 0.011111111 | 0.0034172229 | 1.701107 | AGUGUG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| HNRNPH2 | 11 | 9711 | 0.044444444 | 0.0140746688 | 1.658902 | AAGGGA,AAUGUG,AGGGAA,CUGGGG,GAAUGU,GGAAUG,GGAGGA,GGGAAU,GGGGAA,GGGGGA,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| PPIE | 21 | 18315 | 0.081481481 | 0.0265436196 | 1.618107 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAUAAA,AAUAUA,AUAAAU,AUAUAU,AUAUUU,AUUAUU,AUUUAA,UAUAUU,UAUUUA,UUAAAU,UUUAAA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS3 | 3 | 3429 | 0.014814815 | 0.0049707696 | 1.575499 | AAAUAA,AAUAAA,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPF | 11 | 10561 | 0.044444444 | 0.0153064921 | 1.537859 | AAGGGA,AAUGUG,AGGGAA,AUGGGA,CUGGGG,GAAUGU,GAGGAA,GGAAUG,GGAGGA,GGGAAU,GGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 11 | 10717 | 0.044444444 | 0.0155325680 | 1.516707 | AAGGGA,AAUGUG,AGGGAA,CUGGGG,GAAUGU,GAGGAA,GGAAUG,GGAGGA,GGGAAU,GGGGAA,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| KHDRBS2 | 1 | 1858 | 0.007407407 | 0.0026940701 | 1.459181 | AAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPH3 | 7 | 7688 | 0.029629630 | 0.0111429292 | 1.410912 | AAGGGA,AAUGUG,AGGGAA,GAAUGU,GGAAUG,GGAGGA,GGGAAU | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| RBM5 | 6 | 6879 | 0.025925926 | 0.0099705232 | 1.378654 | AAAAAA,AGGGAA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| CELF6 | 2 | 2997 | 0.011111111 | 0.0043447134 | 1.354670 | UGUGAG,UGUGAU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM3 | 1 | 2152 | 0.007407407 | 0.0031201361 | 1.247360 | AAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| LIN28A | 3 | 4315 | 0.014814815 | 0.0062547643 | 1.244013 | AGGAGA,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ZFP36 | 25 | 28400 | 0.096296296 | 0.0411588414 | 1.226278 | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAAGC,AAAAUA,AAAGGA,AAAUAA,AAGCAA,AAUAAA,AGCAAA,AGGAAA,AUAAAU,AUUUAA,GGAAAG,UAAACA,UAUUUA,UUUAAA | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| RBM46 | 3 | 4554 | 0.014814815 | 0.0066011240 | 1.166257 | AAUGAA,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CELF1 | 1 | 2391 | 0.007407407 | 0.0034664959 | 1.095491 | CUGUCU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPC5 | 1 | 2400 | 0.007407407 | 0.0034795387 | 1.090073 | GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
RBP on BSJ (Exon Only)
Plot
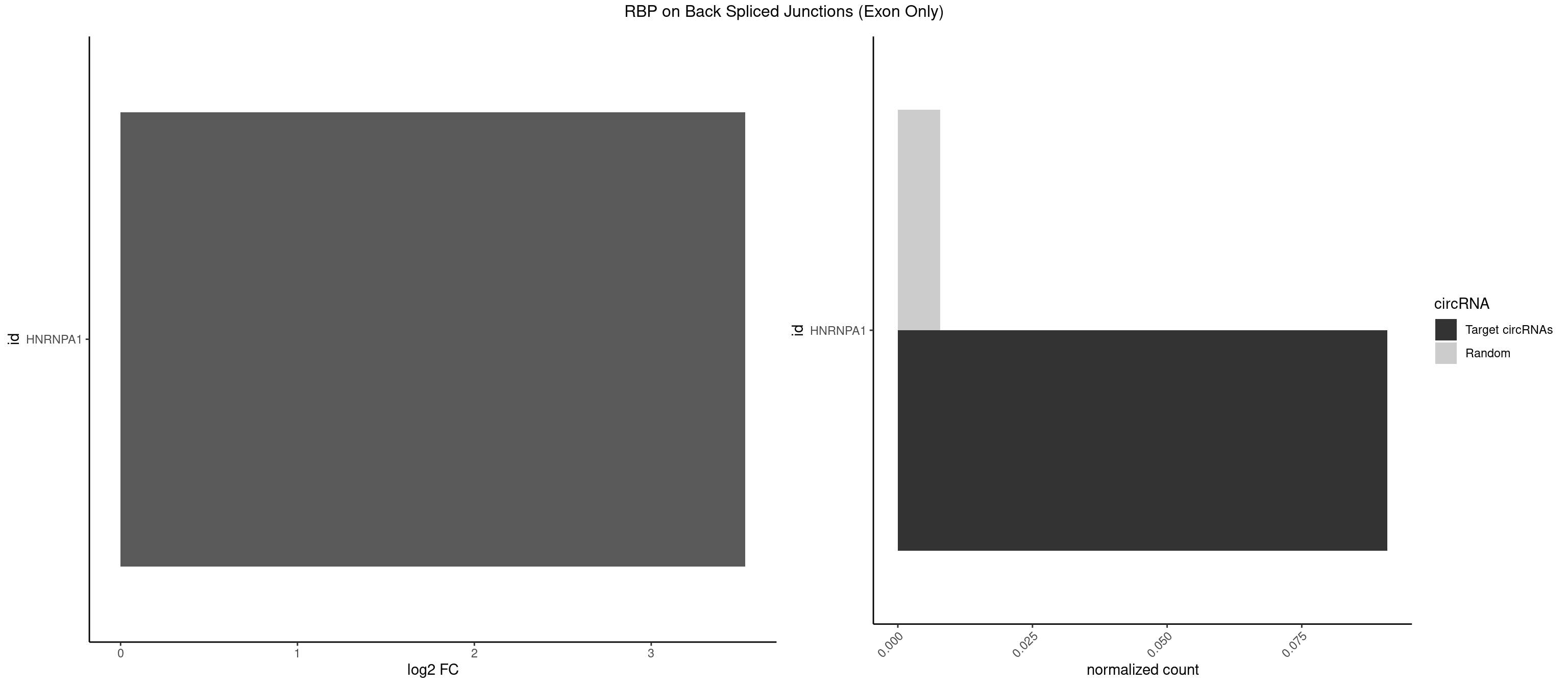
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | UAGGUC | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
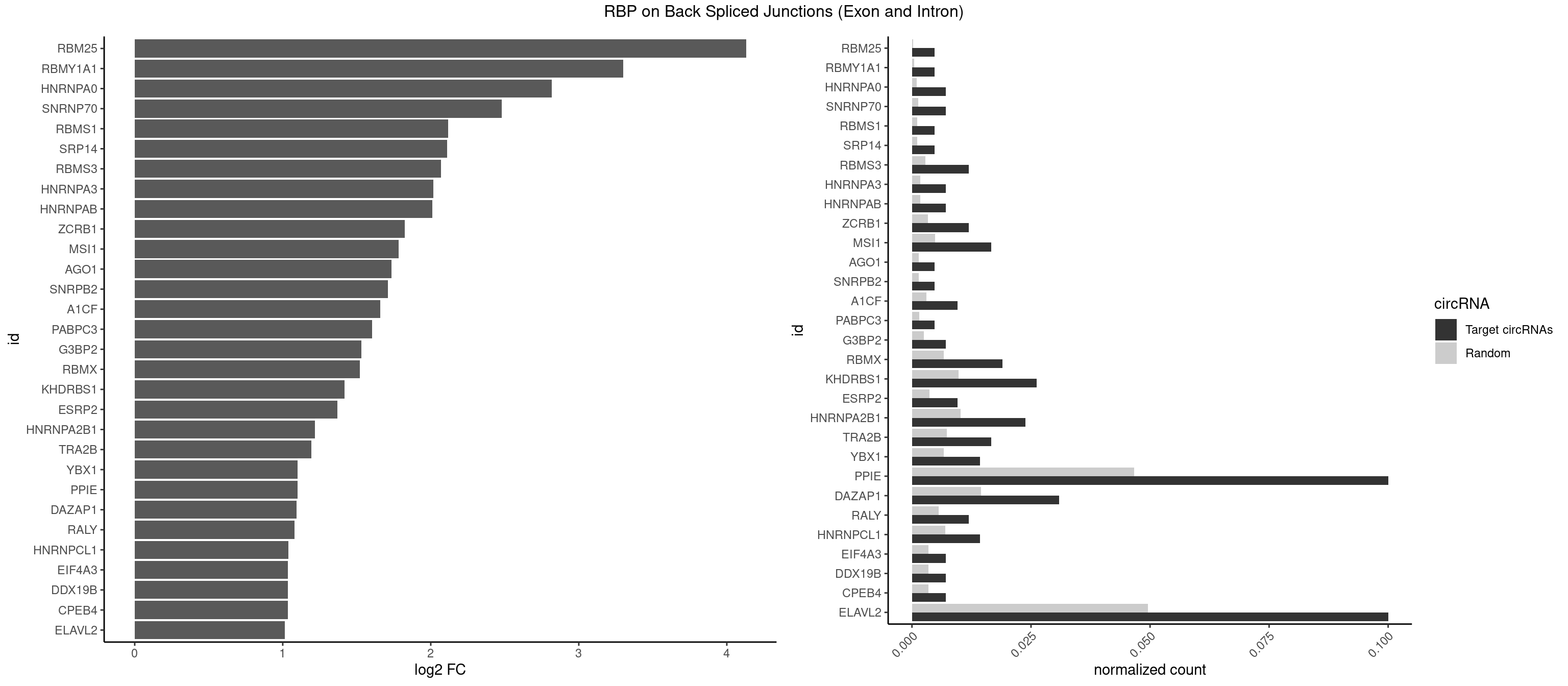
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,AUAUAG,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CAAGGA,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAGACA,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | ACUUAA,AUUUAA,GCUUAA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| MSI1 | 6 | 961 | 0.016666667 | 0.0048468360 | 1.781850 | AGGAAG,AGGAGG,AGGUAG,AGGUGG,UAGGUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | GAUCAG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUU,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBMX | 7 | 1316 | 0.019047619 | 0.0066354293 | 1.521349 | AAGUAA,AGGAAG,AGUAAC,AGUGUU,AUCCCA,GGAAGG,UAAGAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| KHDRBS1 | 10 | 1945 | 0.026190476 | 0.0098045143 | 1.417524 | AUAAAA,AUUUAA,CUAAAA,UAAAAA,UAAAAC,UUAAAA,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGAAG,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPA2B1 | 9 | 2033 | 0.023809524 | 0.0102478839 | 1.216213 | AAUUUA,AUAGCA,AUUUAA,CAAGGA,GAAGCC,GCCAAG,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAGAAC,AAGAAU,AGGAAG,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | CACACC,CACCAC,CCACCA,CCAGCA,CCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| PPIE | 41 | 9262 | 0.100000000 | 0.0466696896 | 1.099442 | AAAAAU,AAAAUA,AAAAUU,AAAUAU,AAAUUA,AAUAUA,AAUUUA,AAUUUU,AUAAAA,AUAUAA,AUUAUA,AUUUAA,AUUUUA,UAAAAA,UAAUUU,UAUAAA,UAUUAU,UAUUUU,UUAAAA,UUAAUU,UUAUAA,UUAUUA,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| DAZAP1 | 12 | 2878 | 0.030952381 | 0.0145052398 | 1.093476 | AAUUUA,AGGAAG,AGGUAA,AGGUAG,AGUUAA,GUAACG,UAGGUA,UAGUUU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| ELAVL2 | 41 | 9826 | 0.100000000 | 0.0495112858 | 1.014171 | AAUUUA,AAUUUU,AUUUAA,AUUUAG,AUUUUA,CUUUUA,CUUUUU,GUAUUG,GUUUUA,UAAUUU,UAUUAU,UAUUGA,UAUUUU,UCUUUU,UGUAUU,UUAAGU,UUAAUU,UUAUAC,UUAUUG,UUCUUU,UUUAAG,UUUAGU,UUUAUA,UUUAUU,UUUCUU,UUUUAA,UUUUAU,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.