circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000140374:-:15:76274411:76292700
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000140374:-:15:76274411:76292700 | ENSG00000140374 | ENST00000688154 | - | 15 | 76274412 | 76292700 | 630 | GUGGCACAAGAUCUCUGUAAAGUAGCAGGCAUAGCAAAAGUUCUGGUGGCUCAGCAUGAUGUGUACAAAGGCCUACUUCCAGAGGAACUGACACCAUUGAUUUUGGCAACUCAGAAGCAGUUCAAUUACACACACAUCUGUGCUGGAGCAUCUGCCUUCGGAAAGAACCUUUUGCCCAGAGUAGCAGCCAAACUUGAGGUUGCCCCGAUUUCUGACAUCAUUGCAAUCAAGUCACCUGACACAUUUGUGAGAACUAUUUAUGCAGGAAAUGCUCUAUGUACAGUGAAGUGUGAUGAGAAAGUGAAAGUGUUUUCUGUCCGUGGAACAUCCUUUGAUGCUGCAGCAACAAGUGGCGGUAGUGCCAGUUCAGAAAAGGCAUCAAGUACUUCACCAGUGGAAAUAUCAGAGUGGCUUGACCAGAAAUUAACAAAAAGUGAUCGACCAGAGCUAACAGGUGCCAAAGUGGUGGUAUCUGGUGGUCGAGGCUUGAAGAGUGGAGAGAACUUUAAGUUGUUAUAUGACUUGGCAGAUCAACUACAUGCUGCAGUUGGUGCUUCCCGUGCUGCUGUUGAUGCUGGCUUUGUUCCCAAUGACAUGCAAGUUGGACAGACGGGAAAAAUAGUAGCACCAGUGGCACAAGAUCUCUGUAAAGUAGCAGGCAUAGCAAAAGUUCUGGUGGC | circ |
| ENSG00000140374:-:15:76274411:76292700 | ENSG00000140374 | ENST00000688154 | - | 15 | 76274412 | 76292700 | 22 | UAGUAGCACCAGUGGCACAAGA | bsj |
| ENSG00000140374:-:15:76274411:76292700 | ENSG00000140374 | ENST00000688154 | - | 15 | 76292691 | 76292900 | 210 | CUCUCAGAGUGCUGGGAUUACAGGCUUGAGCCACUGCGCCCGGCCGGAAUAUAUUUUAAUUCUUAACUGGCCCUUUUGCUAAUAAGAAUAAUUUUUACAGUUGGCCAUUAAGAUUUUAUGAUCUGUUGAUAUUUAUAGUGGUGGCUUUAUUUGUAUAUUUCUGAUAGAUGGAUAAUUAGGAACUUAUCUUUUAAUCAUAGGUGGCACAAG | ie_up |
| ENSG00000140374:-:15:76274411:76292700 | ENSG00000140374 | ENST00000688154 | - | 15 | 76274212 | 76274421 | 210 | AGUAGCACCAGUAAGUAUUGCAAUAAAAUAUGCUGUGUAAAAUGUUAUGGGGAUAAGUACUGUAUGUAUUUGUUGAACAGUGAUCUCAGUGAUUUACCUUACUCAGUGUUCCUGAGCAGUUAGCCAGUACCAUGGUGAAUAUGUCAUUUUCACAAGGUCUGUGUAGUUCUUGAUCCUAGUAAAAUGCCCAGCAGUUCUUAAAUCCAGCUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
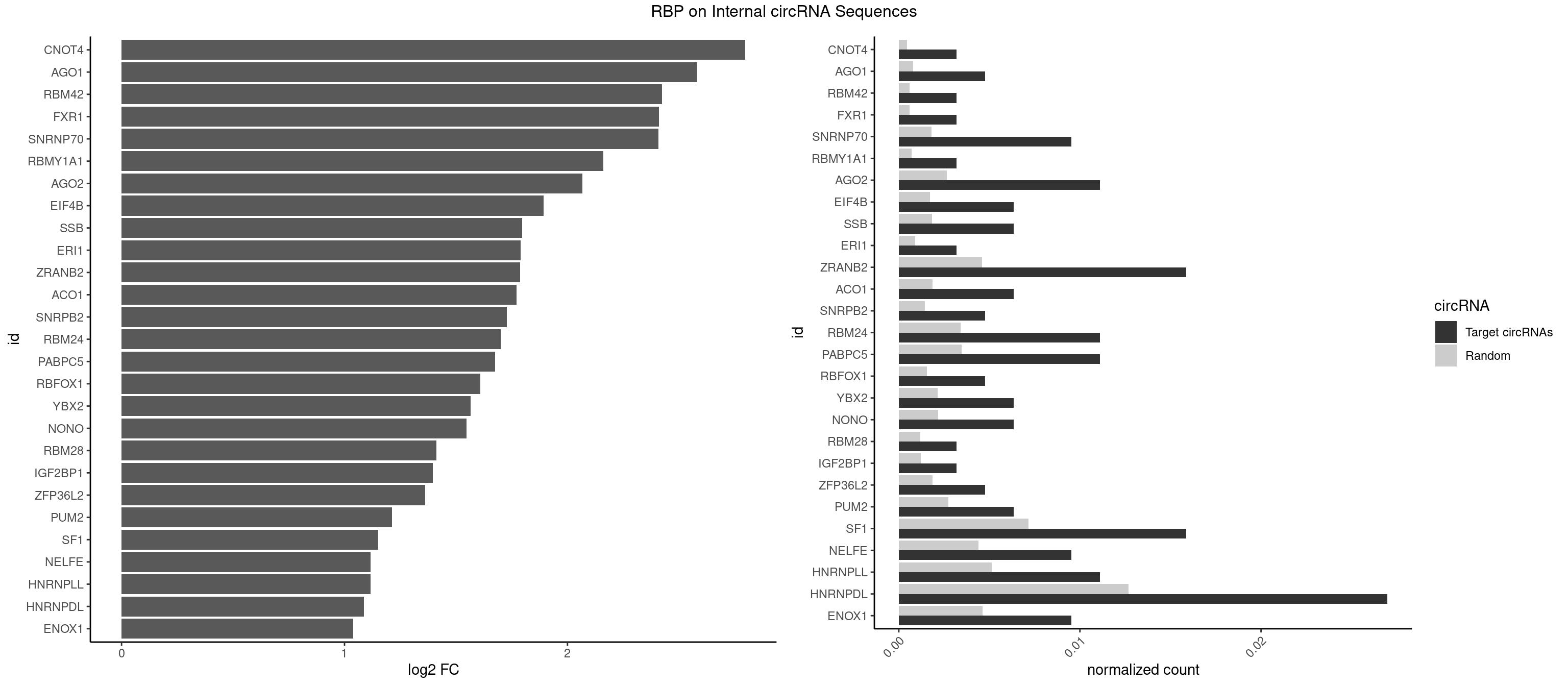
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.003174603 | 0.0004564992 | 2.797892 | GACAGA | GACAGA |
| AGO1 | 2 | 548 | 0.004761905 | 0.0007956130 | 2.581400 | GGUAGU,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBM42 | 1 | 407 | 0.003174603 | 0.0005912752 | 2.424675 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.003174603 | 0.0005970720 | 2.410599 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SNRNP70 | 5 | 1237 | 0.009523810 | 0.0017941145 | 2.408267 | AAUCAA,AUCAAG,GAUCAA,GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMY1A1 | 1 | 489 | 0.003174603 | 0.0007101099 | 2.160462 | ACAAGA | ACAAGA,CAAGAC |
| AGO2 | 6 | 1830 | 0.011111111 | 0.0026534924 | 2.066039 | AAAGUG,GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| EIF4B | 3 | 1179 | 0.006349206 | 0.0017100607 | 1.892529 | GUUGGA,UCGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SSB | 3 | 1260 | 0.006349206 | 0.0018274462 | 1.796747 | GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ERI1 | 1 | 632 | 0.003174603 | 0.0009173461 | 1.791038 | UUCAGA | UUCAGA,UUUCAG |
| ZRANB2 | 9 | 3173 | 0.015873016 | 0.0045997733 | 1.786942 | CGGUAG,GAGGUU,GGUGGU,GUAAAG,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ACO1 | 3 | 1283 | 0.006349206 | 0.0018607779 | 1.770670 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SNRPB2 | 2 | 991 | 0.004761905 | 0.0014376103 | 1.727866 | AUUGCA,UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM24 | 6 | 2357 | 0.011111111 | 0.0034172229 | 1.701107 | AGAGUG,AGUGUG,GAGUGG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPC5 | 6 | 2400 | 0.011111111 | 0.0034795387 | 1.675035 | AGAAAA,AGAAAG,AGAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBFOX1 | 2 | 1077 | 0.004761905 | 0.0015622419 | 1.607921 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| YBX2 | 3 | 1480 | 0.006349206 | 0.0021462711 | 1.564744 | AACAUC,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 3 | 1498 | 0.006349206 | 0.0021723567 | 1.547315 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM28 | 1 | 822 | 0.003174603 | 0.0011926949 | 1.412351 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGF2BP1 | 1 | 831 | 0.003174603 | 0.0012057377 | 1.396660 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ZFP36L2 | 2 | 1277 | 0.004761905 | 0.0018520827 | 1.362390 | AUUUAU,UAUUUA | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PUM2 | 3 | 1890 | 0.006349206 | 0.0027404447 | 1.212166 | UGUAAA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SF1 | 9 | 4931 | 0.015873016 | 0.0071474739 | 1.151071 | ACAGAC,ACUGAC,AUUAAC,CUAACA,GCUAAC,UAACAA,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| NELFE | 5 | 3028 | 0.009523810 | 0.0043896388 | 1.117437 | CUGGCU,GCUAAC,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| HNRNPLL | 6 | 3534 | 0.011111111 | 0.0051229360 | 1.116960 | ACACAC,ACACCA,CACACA,CAGACG | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPDL | 16 | 8759 | 0.026984127 | 0.0126950266 | 1.087848 | AACCUU,ACACCA,ACCAGA,ACUUUA,AGUAGC,CACCAG,CUAACA,CUUUAA,GAACUA,GAGUAG,UAACAG,UCUGAC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| ENOX1 | 5 | 3195 | 0.009523810 | 0.0046316558 | 1.040011 | GGACAG,GUACAG,UGGACA,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
RBP on BSJ (Exon Only)
Plot
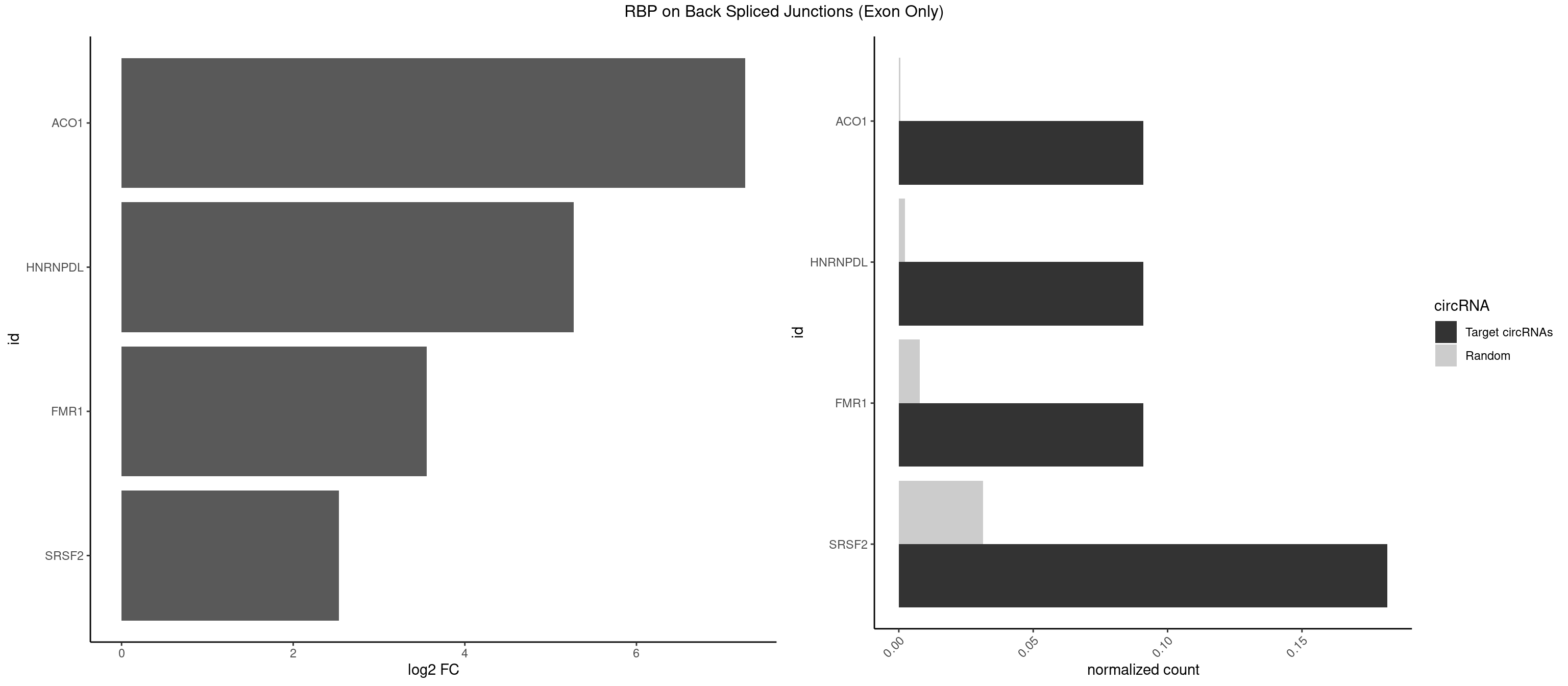
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ACO1 | 1 | 8 | 0.09090909 | 0.0005894682 | 7.268867 | CAGUGG | CAGUGA,CAGUGC,CAGUGG |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | CACCAG | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | AGUGGC | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF2 | 3 | 479 | 0.18181818 | 0.0314383023 | 2.531901 | ACCAGU,CAGUGG,CCAGUG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
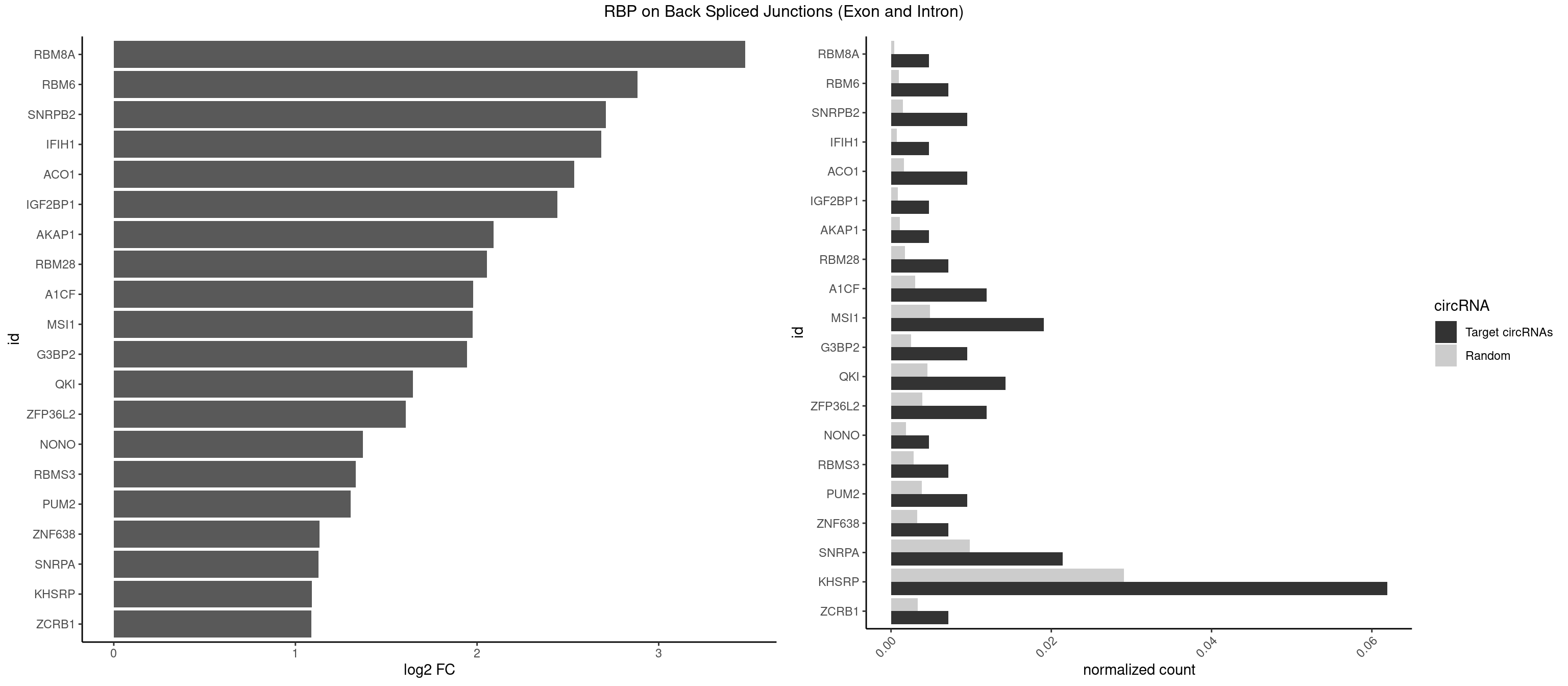
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SNRPB2 | 3 | 288 | 0.009523810 | 0.0014560661 | 2.709463 | AUUGCA,GUAUUG,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ACO1 | 3 | 325 | 0.009523810 | 0.0016424829 | 2.535660 | CAGUGA,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | GUGUAG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| MSI1 | 7 | 961 | 0.019047619 | 0.0048468360 | 1.974496 | AGGUGG,AGUAAG,AGUUAG,AGUUGG,UAGGAA,UAGGUG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | GGAUAA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AAUCAU,ACUUAU,AUCAUA,UAAUCA,UACUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AAUAUA,AUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAUAU,UGUAAA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | AGCAGU,AUGCUG,AUUGCA,GAGCAG,UAUGCU,UUACCU,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| KHSRP | 25 | 5764 | 0.061904762 | 0.0290457477 | 1.091723 | AUAUUU,AUGUAU,AUUUAU,AUUUUA,CAGCUU,CUGUAU,CUGUGU,GUAUAU,GUAUGU,UAUAUU,UAUUUA,UAUUUU,UGUAUA,UGUAUG,UGUGUA,UUAUUU,UUGUAU,UUUAUA,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUUA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.