circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000150403:+:13:113539356:113539507
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000150403:+:13:113539356:113539507 | ENSG00000150403 | ENST00000434316 | + | 13 | 113539357 | 113539507 | 151 | GUCACGGAGCUGCUGGACGUCUCCAUGGAGCUGGGCUGUUUCCUGGCUGGAGCGCUCGUCUCCUCUCAGGGCCCCGUGGUCACCGAGGAGAUCGCCACCUCCAUCGAACCCAUCCGCGACUUCCUGGCCAUCGUUUUCUUCGCCUCCAUAGGUCACGGAGCUGCUGGACGUCUCCAUGGAGCUGGGCUGUUUCCUGGCUGG | circ |
| ENSG00000150403:+:13:113539356:113539507 | ENSG00000150403 | ENST00000434316 | + | 13 | 113539357 | 113539507 | 22 | CGCCUCCAUAGGUCACGGAGCU | bsj |
| ENSG00000150403:+:13:113539356:113539507 | ENSG00000150403 | ENST00000434316 | + | 13 | 113539157 | 113539366 | 210 | GUUAAUUCAACUAAUAGCAGCCUCCACAUGUCACCUCUCAGACCCAGCAGACGACCCAGGACCAGACACACACACGCUCUGUUUGUGCAGCUUCUGUGUUUUGUCGUGGUGGCUCUGCUGGGUCUCGGGGUGGCCUUGGGAUGGGGGUGACCCUGGUGCACUGUGGGGUCUCAUGUAAAGCUGCGUCCUGCCUCUCUCAGGUCACGGAGC | ie_up |
| ENSG00000150403:+:13:113539356:113539507 | ENSG00000150403 | ENST00000434316 | + | 13 | 113539498 | 113539707 | 210 | GCCUCCAUAGGUAAUCUUCCUUCUUUACAUUAUGAUUGUGUUGGUUAAACGUAUGCAGAGUGGUUCUGUAAUAUGUUUACGUGCAUAUAUAUUUAUAUAGCAAAAAUGGUUAUCUUAAAAAUUAAGUGUAUUCAGCAUAUUUCAAUUUGGAUUGCUAGUAGCCAGAAACUGUUUUCUGCUUUAUAGAGCGCAGAGAAAUCUUAUUUUAUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
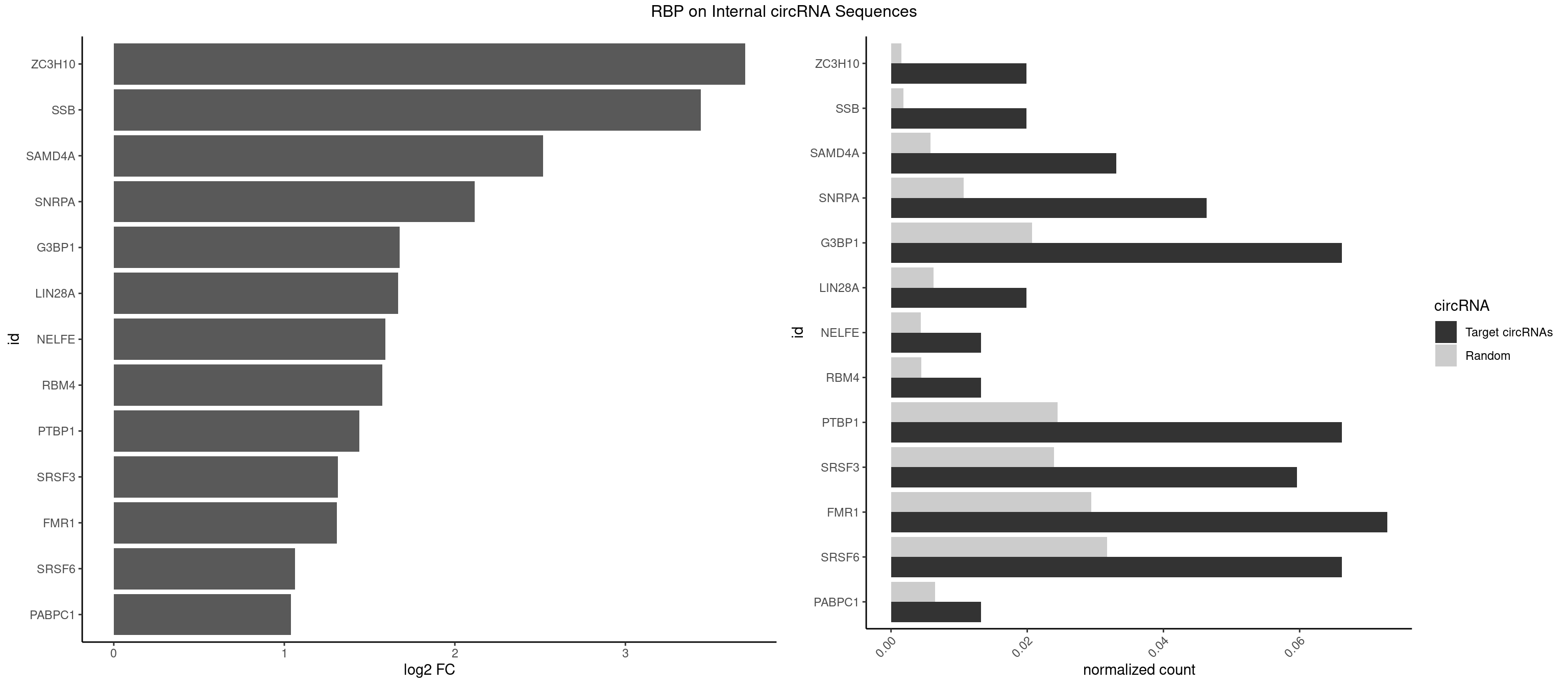
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 2 | 1053 | 0.01986755 | 0.001527461 | 3.701207 | GAGCGC,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SSB | 2 | 1260 | 0.01986755 | 0.001827446 | 3.442513 | CUGUUU,GCUGUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SAMD4A | 4 | 3992 | 0.03311258 | 0.005786671 | 2.516574 | CUGGAC,CUGGCC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SNRPA | 6 | 7380 | 0.04635762 | 0.010696574 | 2.115658 | AGGAGA,GGAGAU,GUUUCC,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| G3BP1 | 9 | 14282 | 0.06622517 | 0.020698980 | 1.677820 | ACCCAU,AUCCGC,CAUAGG,CAUCCG,CCAUAG,CCAUCC,CCAUCG,CCCAUC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| LIN28A | 2 | 4315 | 0.01986755 | 0.006254764 | 1.667387 | AGGAGA,GGAGAU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NELFE | 1 | 3028 | 0.01324503 | 0.004389639 | 1.593277 | CUGGCU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 1 | 3065 | 0.01324503 | 0.004443259 | 1.575761 | CUUCCU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PTBP1 | 9 | 16854 | 0.06622517 | 0.024426333 | 1.438942 | CUCCAU,CUUCCU,UCCAUC,UCCUCU,UUCUUC,UUUCUU,UUUUCU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| SRSF3 | 8 | 16536 | 0.05960265 | 0.023965486 | 1.314418 | AUCGUU,CAUCGA,CAUCGU,CUCGUC,GACUUC,UCUCCA,UCUUCG,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| FMR1 | 10 | 20291 | 0.07284768 | 0.029407247 | 1.308711 | AGCUGG,AUGGAG,CAUAGG,CUGGGC,GCGACU,GCUGGG,GGCUGG,GGGCUG,UGGCUG | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| SRSF6 | 9 | 21880 | 0.06622517 | 0.031710032 | 1.062440 | CACGGA,CCUCUC,CCUGGC,CUCAGG,GACGUC,UCCUGG,UCUCAG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| PABPC1 | 1 | 4443 | 0.01324503 | 0.006440262 | 1.040260 | CGAACC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
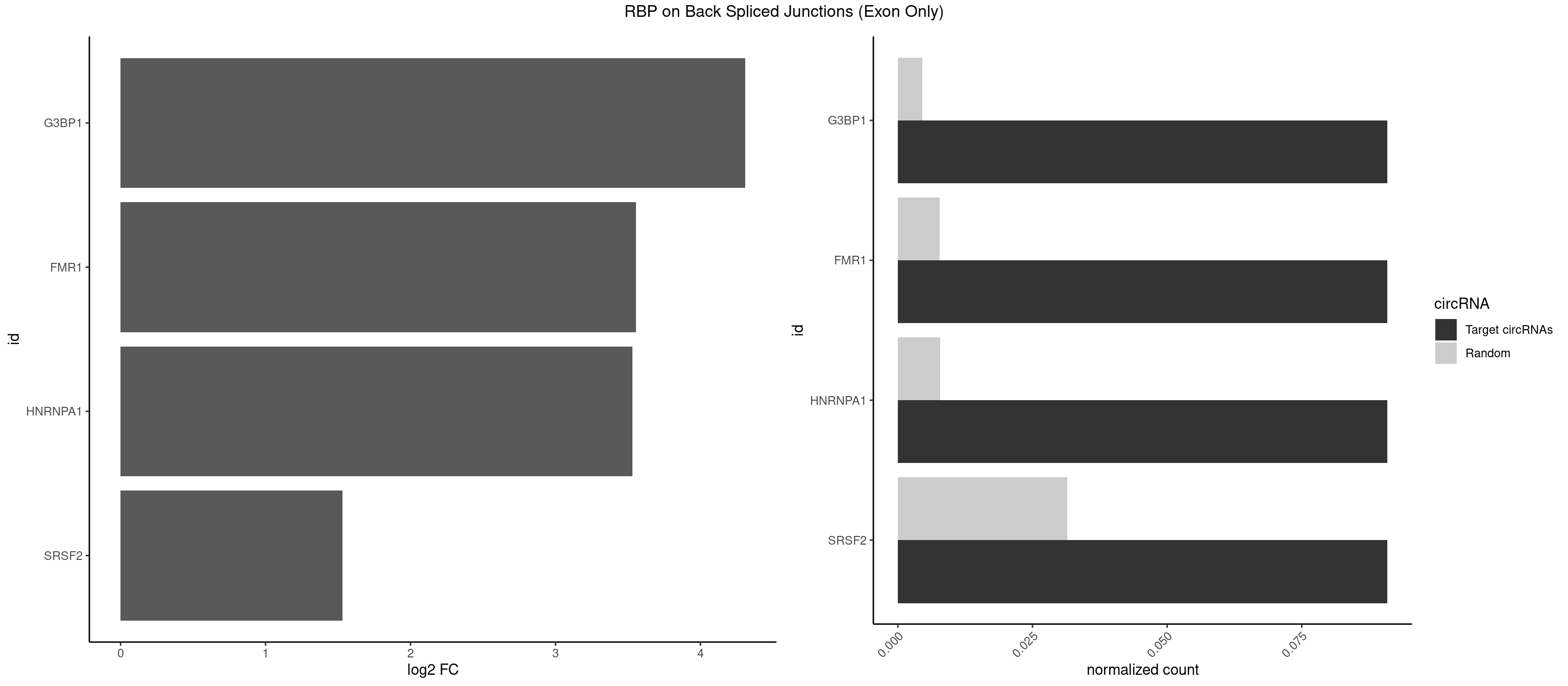
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| G3BP1 | 1 | 69 | 0.09090909 | 0.004584752 | 4.309509 | CAUAGG | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | CAUAGG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | UAGGUC | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | GGUCAC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
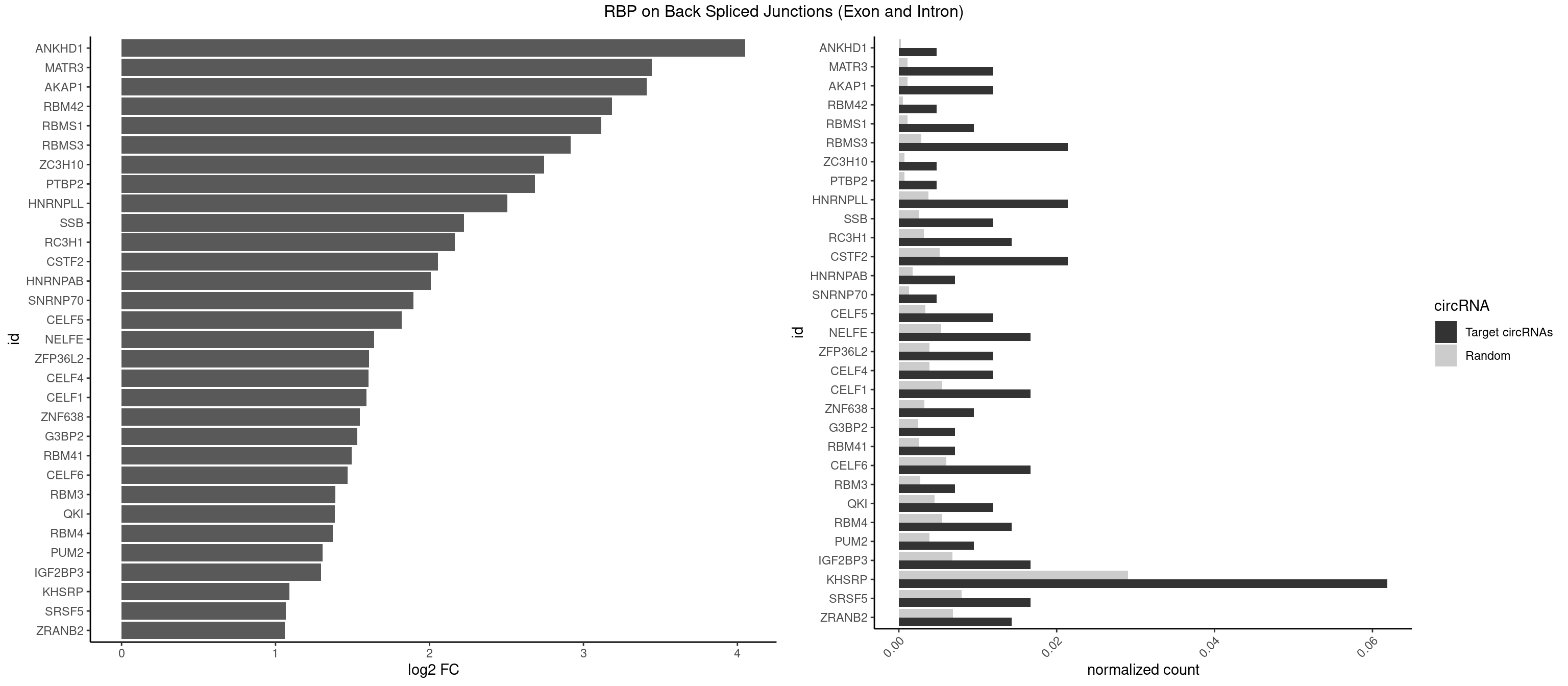
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| MATR3 | 4 | 216 | 0.011904762 | 0.0010933091 | 3.444765 | AAUCUU,AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| AKAP1 | 4 | 221 | 0.011904762 | 0.0011185006 | 3.411901 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 | 8 | 562 | 0.021428571 | 0.0028365578 | 2.917322 | AUAUAG,AUAUAU,CAUAUA,UAUAGA,UAUAGC,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| HNRNPLL | 8 | 748 | 0.021428571 | 0.0037736800 | 2.505492 | ACACAC,AGACGA,CACACA,CAGACG,GACGAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RC3H1 | 5 | 631 | 0.014285714 | 0.0031841999 | 2.165570 | CCUUCU,CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CSTF2 | 8 | 1022 | 0.021428571 | 0.0051541717 | 2.055723 | GUGUUG,GUGUUU,GUUUUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUUG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUCU,GGUCUC,UCUCUC,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUUG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | GUGUUG,GUUUGU,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUCU,GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CELF6 | 6 | 1196 | 0.016666667 | 0.0060308343 | 1.466536 | GUGGGG,GUGGUG,GUGUUG,UGUGGG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AGACGA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACACAC,ACUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM4 | 5 | 1094 | 0.014285714 | 0.0055169287 | 1.372636 | CCUUCU,CUUCCU,CUUCUU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAUAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| IGF2BP3 | 6 | 1348 | 0.016666667 | 0.0067966546 | 1.294069 | AAUUCA,ACACAC,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| KHSRP | 25 | 5764 | 0.061904762 | 0.0290457477 | 1.091723 | AGCCUC,AGCUUC,AUAUUU,AUUUAU,AUUUUA,CAGCCU,CAGCUU,CUGCCU,CUGCUU,CUGUGU,GCCUCC,GUGUAU,UAGGUA,UAUAUU,UAUUUA,UAUUUU,UCCUUC,UGCCUC,UUAUUU,UUGUGU,UUUAUA,UUUAUU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | CACGGA,CGCAGA,UGCAGA,UGCAGC,UGCAUA,UGCGUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AGGUAA,GGGUGA,GUAAAG,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.