circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000140391:-:15:77052384:77056255
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000140391:-:15:77052384:77056255 | ENSG00000140391 | ENST00000267970 | - | 15 | 77052385 | 77056255 | 606 | GGGGCAGCUGGCAUUUUAUGCUAUGUGGGAGCCUAUGUCUUCAUCACUUAUGAUGACUAUGACCACUUCUUUGAAGAUGUGUACACGCUCAUCCCUGCUGUAGUGAUCAUAGCUGUAGGAGCCCUGCUUUUCAUCAUUGGGCUAAUUGGCUGCUGUGCCACAAUCCGGGAAAGUCGCUGUGGACUUGCCACGUUUGUCAUCAUCCUGCUCUUGGUUUUUGUCACAGAAGUUGUUGUAGUGGUUUUGGGAUAUGUUUACAGAGCAAAGGUGGAAAAUGAGGUUGAUCGCAGCAUUCAGAAAGUGUAUAAGACCUACAAUGGAACCAACCCUGAUGCUGCUAGCCGGGCUAUUGAUUAUGUACAGAGACAGCUGCAUUGUUGUGGAAUUCACAACUACUCAGACUGGGAAAAUACAGAUUGGUUCAAAGAAACCAAAAACCAGAGUGUCCCUCUUAGCUGCUGCAGAGAGACUGCCAGCAAUUGUAAUGGCAGCCUGGCCCACCCUUCCGACCUCUAUGCUGAGGGGUGUGAGGCUCUAGUAGUGAAGAAGCUACAAGAAAUCAUGAUGCAUGUGAUCUGGGCCGCACUGGCAUUUGCAGCUAUUCAGGGGGCAGCUGGCAUUUUAUGCUAUGUGGGAGCCUAUGUCUUCAUCACUUA | circ |
| ENSG00000140391:-:15:77052384:77056255 | ENSG00000140391 | ENST00000267970 | - | 15 | 77052385 | 77056255 | 22 | CAGCUAUUCAGGGGGCAGCUGG | bsj |
| ENSG00000140391:-:15:77052384:77056255 | ENSG00000140391 | ENST00000267970 | - | 15 | 77056246 | 77056455 | 210 | AAAGUAGGUGAAGUGUGGAUAACUUUUUAAUUUUAUCUAGUAUAGAAAUGCUCUUAACAAGGAAUCAUAGAAUUUUCAGCUUUACUGAAAAAAGUCUAUAACAGUUACGGUAACUCUCAUUAAAUUAUACCAUACUAAGCAAUAAUAGGAAAUUUUUACCAGCAGUGCUUAGAGACCAGUACUGACUUUGUUUUCUACAGGGGGCAGCUG | ie_up |
| ENSG00000140391:-:15:77052384:77056255 | ENSG00000140391 | ENST00000267970 | - | 15 | 77052185 | 77052394 | 210 | AGCUAUUCAGGUAAGCACAGCUUGCCAAGGAGUUCUAUCGGAGUGAGUUGGUUCUCUGUUGUCAGCCCUGAAUGGUUUUAAUCCCCAUCACAGUGAAUCUUUGUGUAUGUAGGUAAUGUGGAAGUGACUGAGGUACUGGCCUCCAGUUUCCCAUGUUUUCCAUUUCUUGUCUAUACACCUAGACCCGGAAUGGUCUAAAGAAACCAGAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
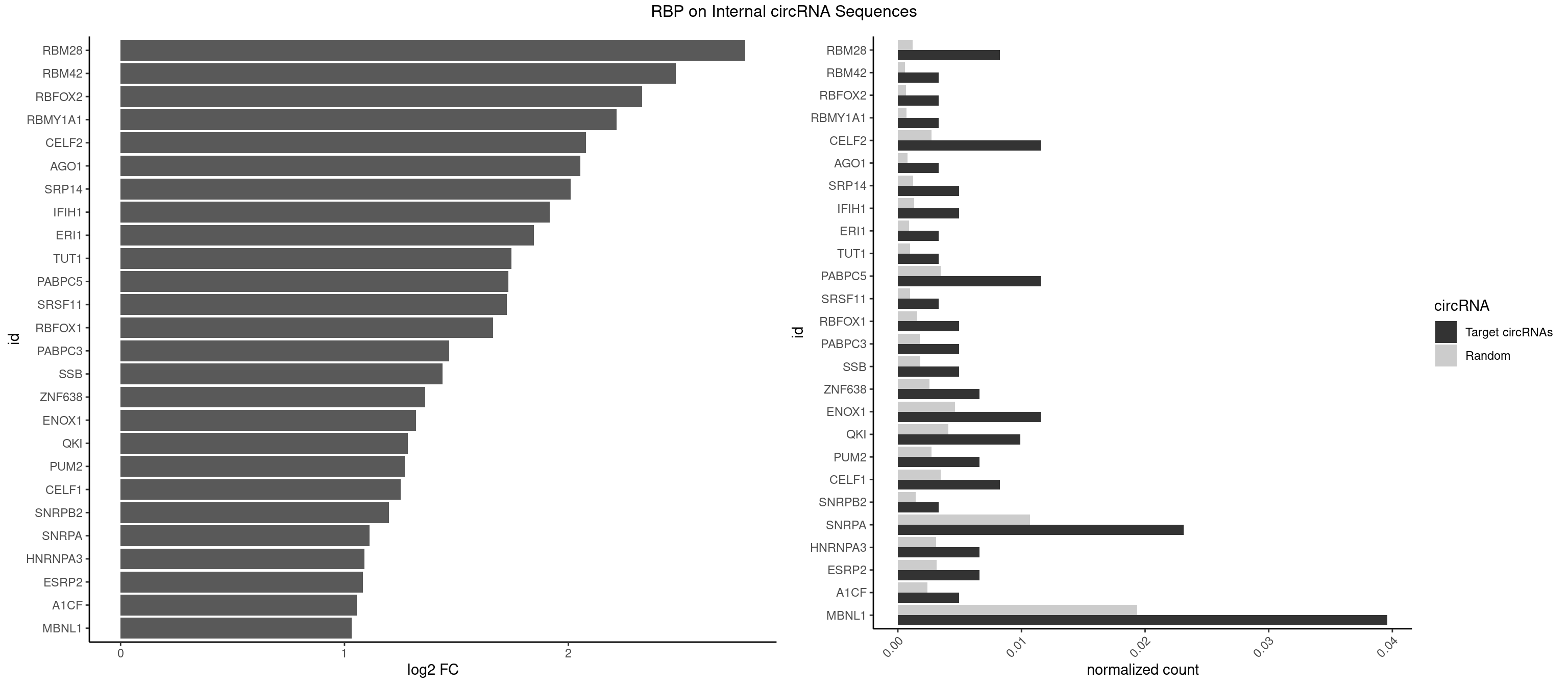
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM28 | 4 | 822 | 0.008250825 | 0.0011926949 | 2.790313 | AGUAGU,UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBM42 | 1 | 407 | 0.003300330 | 0.0005912752 | 2.480709 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBFOX2 | 1 | 452 | 0.003300330 | 0.0006564894 | 2.329767 | UGCAUG | UGACUG,UGCAUG |
| RBMY1A1 | 1 | 489 | 0.003300330 | 0.0007101099 | 2.216496 | ACAAGA | ACAAGA,CAAGAC |
| CELF2 | 6 | 1886 | 0.011551155 | 0.0027346479 | 2.078610 | AUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| AGO1 | 1 | 548 | 0.003300330 | 0.0007956130 | 2.052472 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 847 | 0.004950495 | 0.0012289250 | 2.010176 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 2 | 904 | 0.004950495 | 0.0013115296 | 1.916322 | GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ERI1 | 1 | 632 | 0.003300330 | 0.0009173461 | 1.847072 | UUCAGA | UUCAGA,UUUCAG |
| TUT1 | 1 | 678 | 0.003300330 | 0.0009840095 | 1.745866 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC5 | 6 | 2400 | 0.011551155 | 0.0034795387 | 1.731069 | AGAAAG,AGAAAU,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SRSF11 | 1 | 688 | 0.003300330 | 0.0009985015 | 1.724774 | AAGAAG | AAGAAG |
| RBFOX1 | 2 | 1077 | 0.004950495 | 0.0015622419 | 1.663955 | GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC3 | 2 | 1234 | 0.004950495 | 0.0017897669 | 1.467801 | AAAAAC,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SSB | 2 | 1260 | 0.004950495 | 0.0018274462 | 1.437744 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ZNF638 | 3 | 1773 | 0.006600660 | 0.0025708878 | 1.360344 | GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ENOX1 | 6 | 3195 | 0.011551155 | 0.0046316558 | 1.318437 | AAUACA,AGACAG,AUACAG,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| QKI | 5 | 2805 | 0.009900990 | 0.0040664663 | 1.283797 | AAUCAU,ACUUAU,AUCAUA,CUACUC,UACUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PUM2 | 3 | 1890 | 0.006600660 | 0.0027404447 | 1.268200 | UGUACA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CELF1 | 4 | 2391 | 0.008250825 | 0.0034664959 | 1.251060 | GUUGUG,GUUUGU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SNRPB2 | 1 | 991 | 0.003300330 | 0.0014376103 | 1.198938 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| SNRPA | 13 | 7380 | 0.023102310 | 0.0106965744 | 1.110888 | AGUAGU,AUGCUG,CCUGCU,CUGCUA,GUAGUG,UAUGCU,UCCUGC | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| HNRNPA3 | 3 | 2140 | 0.006600660 | 0.0031027457 | 1.089065 | AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 3 | 2150 | 0.006600660 | 0.0031172377 | 1.082342 | GGGAAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| A1CF | 2 | 1642 | 0.004950495 | 0.0023810421 | 1.055980 | UAAUUG,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| MBNL1 | 23 | 13372 | 0.039603960 | 0.0193802045 | 1.031061 | ACGCUC,CCUGCU,CGCUGU,CUGCCA,CUGCUA,CUGCUC,CUGCUG,CUGCUU,GCUGCU,GCUUUU,UCGCUG,UGCUGC,UGCUGU,UGCUUU,UUGCCA | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
RBP on BSJ (Exon Only)
Plot
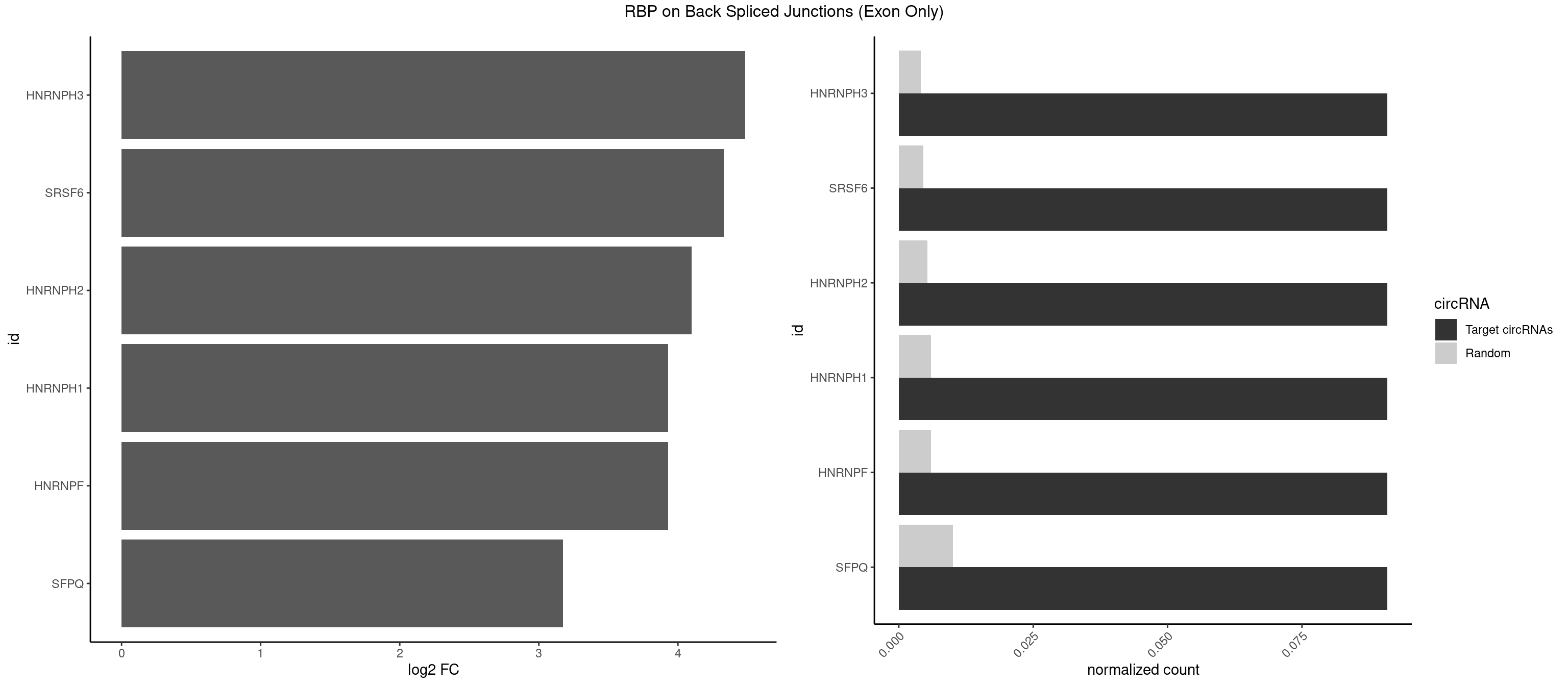
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | GGGGGC | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | UUCAGG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.005305214 | 4.098942 | GGGGGC | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | GGGGGC | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | GGGGGC | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | AGGGGG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
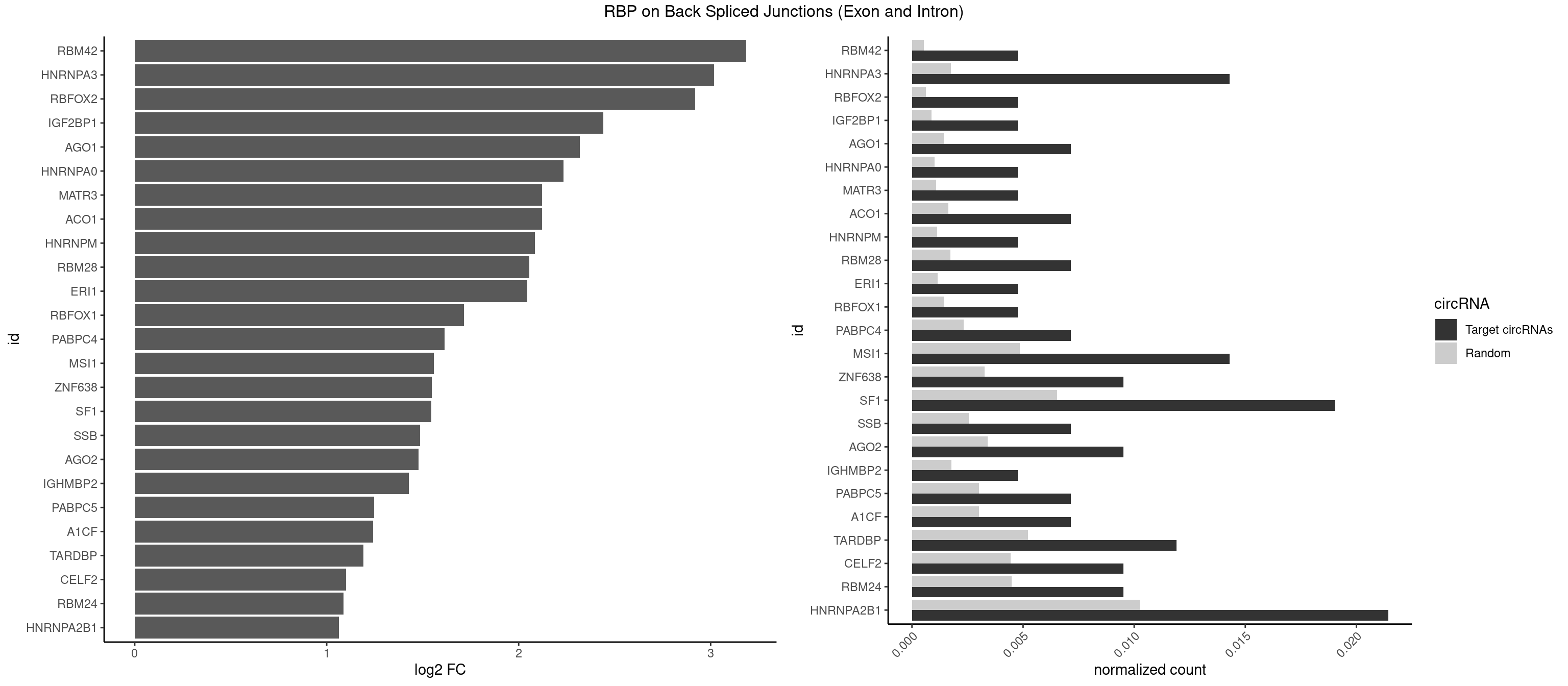
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| HNRNPA3 | 5 | 349 | 0.014285714 | 0.0017634019 | 3.018140 | AAGGAG,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGG,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGUAGG,AGUUGG,UAGGAA,UAGGUA,UAGGUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUCU,GUUGGU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SF1 | 7 | 1294 | 0.019047619 | 0.0065245869 | 1.545652 | ACUAAG,ACUGAC,AUACUA,UAACAA,UACUAA,UACUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| AGO2 | 3 | 677 | 0.009523810 | 0.0034159613 | 1.479247 | AAAAAA,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AGUAUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGG,GUGAAU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | AGUGUG,GAGUGA,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | AAGGAA,AAGGAG,AGUAGG,CAAGGA,CCAAGG,CUAGAC,GCCAAG | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.