circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000106066:-:7:29030576:29096217
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000106066:-:7:29030576:29096217 | ENSG00000106066 | ENST00000409850 | - | 7 | 29030577 | 29096217 | 1032 | AUACAGCCAGAAGAUGCCCCAGUAGUUCUCUGGCUACAGGGUGGGCCGGGAGGUUCAUCCAUGUUUGGACUCUUUGUGGAACAUGGGCCUUAUGUUGUCACAAGUAACAUGACCUUGCGUGACAGAGACUUCCCCUGGACCACAACGCUCUCCAUGCUUUACAUUGACAAUCCAGUGGGCACAGGCUUCAGUUUUACUGAUGAUACCCACGGAUAUGCAGUCAAUGAGGACGAUGUAGCACGGGAUUUAUACAGUGCACUAAUUCAGUUUUUCCAGAUAUUUCCUGAAUAUAAAAAUAAUGACUUUUAUGUCACUGGGGAGUCUUAUGCAGGGAAAUAUGUGCCAGCCAUUGCACACCUCAUCCAUUCCCUCAACCCUGUGAGAGAGGUGAAGAUCAACCUGAACGGAAUUGCUAUUGGAGAUGGAUAUUCUGAUCCCGAAUCAAUUAUAGGGGGCUAUGCAGAAUUCCUGUACCAAAUUGGCUUGUUGGAUGAGAAGCAAAAAAAGUACUUCCAGAAGCAGUGCCAUGAAUGCAUAGAACACAUCAGGAAGCAGAACUGGUUUGAGGCCUUUGAAAUACUGGAUAAACUACUAGAUGGCGACUUAACAAGUGAUCCUUCUUACUUCCAGAAUGUUACAGGAUGUAGUAAUUACUAUAACUUUUUGCGGUGCACGGAACCUGAGGAUCAGCUUUACUAUGUGAAAUUUUUGUCACUCCCAGAGGUGAGACAAGCCAUCCACGUGGGGAAUCAGACUUUUAAUGAUGGAACUAUAGUUGAAAAGUACUUGCGAGAAGAUACAGUACAGUCAGUUAAGCCAUGGUUAACUGAAAUCAUGAAUAAUUAUAAGGUUCUGAUCUACAAUGGCCAACUGGACAUCAUCGUGGCAGCUGCCCUGACAGAGCGCUCCUUGAUGGGCAUGGACUGGAAAGGAUCCCAGGAAUACAAGAAGGCAGAAAAAAAAGUUUGGAAGAUCUUUAAAUCUGACAGUGAAGUGGCUGGUUACAUCCGGCAAGCGGGUGACUUCCAUCAGAUACAGCCAGAAGAUGCCCCAGUAGUUCUCUGGCUACAGGGUGGGCCGGG | circ |
| ENSG00000106066:-:7:29030576:29096217 | ENSG00000106066 | ENST00000409850 | - | 7 | 29030577 | 29096217 | 22 | ACUUCCAUCAGAUACAGCCAGA | bsj |
| ENSG00000106066:-:7:29030576:29096217 | ENSG00000106066 | ENST00000409850 | - | 7 | 29096208 | 29096417 | 210 | GAGGCACAGCCUCAUUCAAAGAACCAAAGAUGAUCAGUACUUUGCUCAUAGUGGAGAGUGGUUACUGAGGUCUGGGGAGGUUUUGUAGGAUUUAGUCACAGCCCUGUAACACAGAGAGGAUUUGGGUAAGUUUGCUUGUGUGUGUUCUGUAGCCUUAAGUGAGUGUUCUGUGUGGUCACUGGUUUUACUGUUUCCUCUAGAUACAGCCAG | ie_up |
| ENSG00000106066:-:7:29030576:29096217 | ENSG00000106066 | ENST00000409850 | - | 7 | 29030377 | 29030586 | 210 | CUUCCAUCAGGUAGGAAGAUGCUUGGGAGCAGGCAGGUUGUGGGAAUGGGAGAGCGGGAUUGAAAAUAAAGCCUGAAUAGCAAUGAGACAUGGCCGACCUUAAGCAAAGCCAUACAACAGCCAGUGUGUGUCUUCUGUGGCCCUACUUUCCACAGAGAAGAGUGGUCUGAGUUCAGACAUACAUAGGACUAAAUAUAAAACCAGACAAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
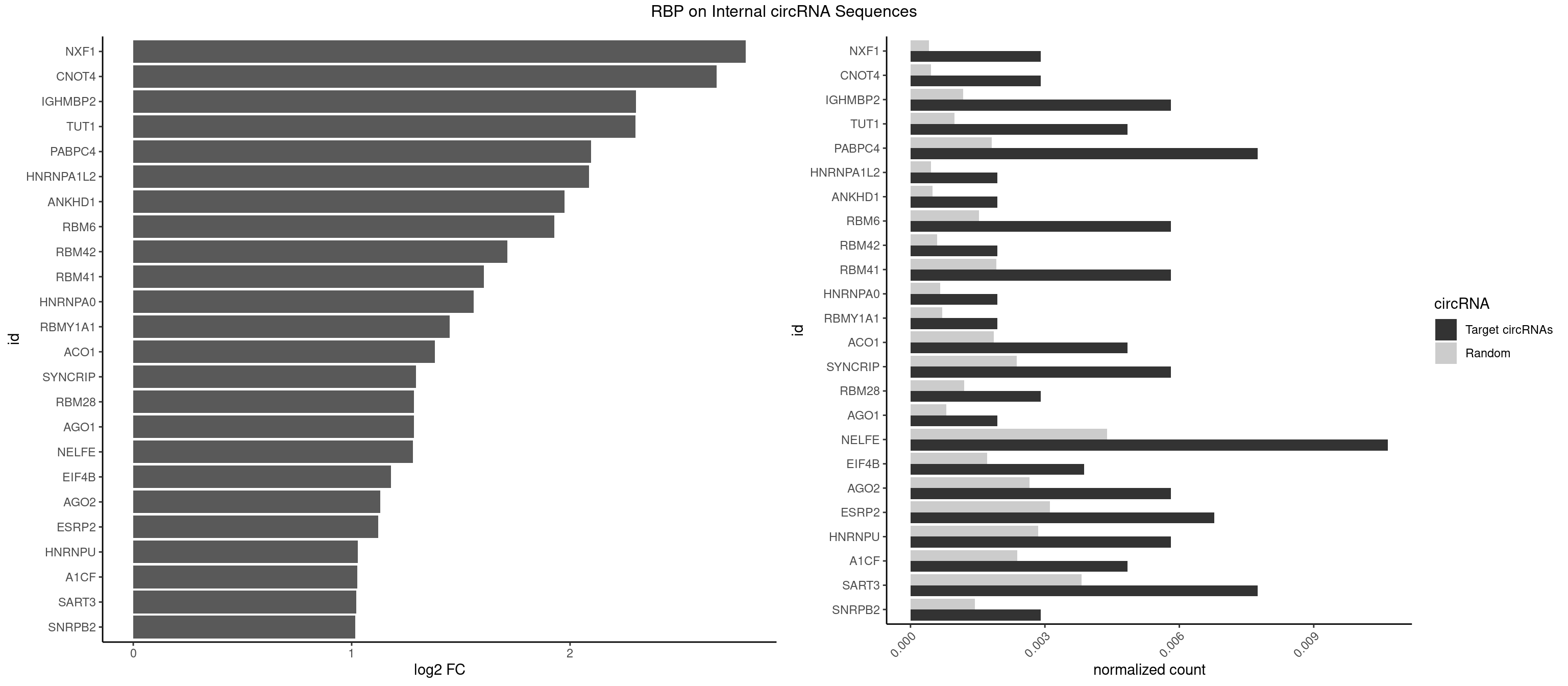
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 2 | 286 | 0.002906977 | 0.0004159215 | 2.805136 | AACCUG | AACCUG |
| CNOT4 | 2 | 314 | 0.002906977 | 0.0004564992 | 2.670835 | GACAGA | GACAGA |
| IGHMBP2 | 5 | 813 | 0.005813953 | 0.0011796520 | 2.301158 | AAAAAA | AAAAAA |
| TUT1 | 4 | 678 | 0.004844961 | 0.0009840095 | 2.299741 | AAAUAC,AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC4 | 7 | 1251 | 0.007751938 | 0.0018144033 | 2.095062 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| HNRNPA1L2 | 1 | 314 | 0.001937984 | 0.0004564992 | 2.085873 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ANKHD1 | 1 | 339 | 0.001937984 | 0.0004927293 | 1.975690 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM6 | 5 | 1054 | 0.005813953 | 0.0015289102 | 1.927016 | AAUCCA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM42 | 1 | 407 | 0.001937984 | 0.0005912752 | 1.712655 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM41 | 5 | 1318 | 0.005813953 | 0.0019115000 | 1.604814 | UACAUU,UACUUG,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPA0 | 1 | 453 | 0.001937984 | 0.0006579386 | 1.558532 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.001937984 | 0.0007101099 | 1.448443 | ACAAGA | ACAAGA,CAAGAC |
| ACO1 | 4 | 1283 | 0.004844961 | 0.0018607779 | 1.380579 | CAGUGA,CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SYNCRIP | 5 | 1634 | 0.005813953 | 0.0023694485 | 1.294968 | AAAAAA | AAAAAA,UUUUUU |
| RBM28 | 2 | 822 | 0.002906977 | 0.0011926949 | 1.285295 | AGUAGU,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| AGO1 | 1 | 548 | 0.001937984 | 0.0007956130 | 1.284418 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| NELFE | 10 | 3028 | 0.010658915 | 0.0043896388 | 1.279886 | CUCUGG,CUGGCU,CUGGUU,UCUCUG,UCUGGC,UGGCUA,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| EIF4B | 3 | 1179 | 0.003875969 | 0.0017100607 | 1.180510 | GUUGGA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| AGO2 | 5 | 1830 | 0.005813953 | 0.0026534924 | 1.131627 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ESRP2 | 6 | 2150 | 0.006782946 | 0.0031172377 | 1.121644 | GGGAAA,GGGAAU,GGGGAA,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPU | 5 | 1965 | 0.005813953 | 0.0028491350 | 1.028996 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| A1CF | 4 | 1642 | 0.004844961 | 0.0023810421 | 1.024892 | AUAAUU,GAUCAG,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SART3 | 7 | 2634 | 0.007751938 | 0.0038186524 | 1.021493 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SNRPB2 | 2 | 991 | 0.002906977 | 0.0014376103 | 1.015847 | AUUGCA,UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
RBP on BSJ (Exon Only)
Plot
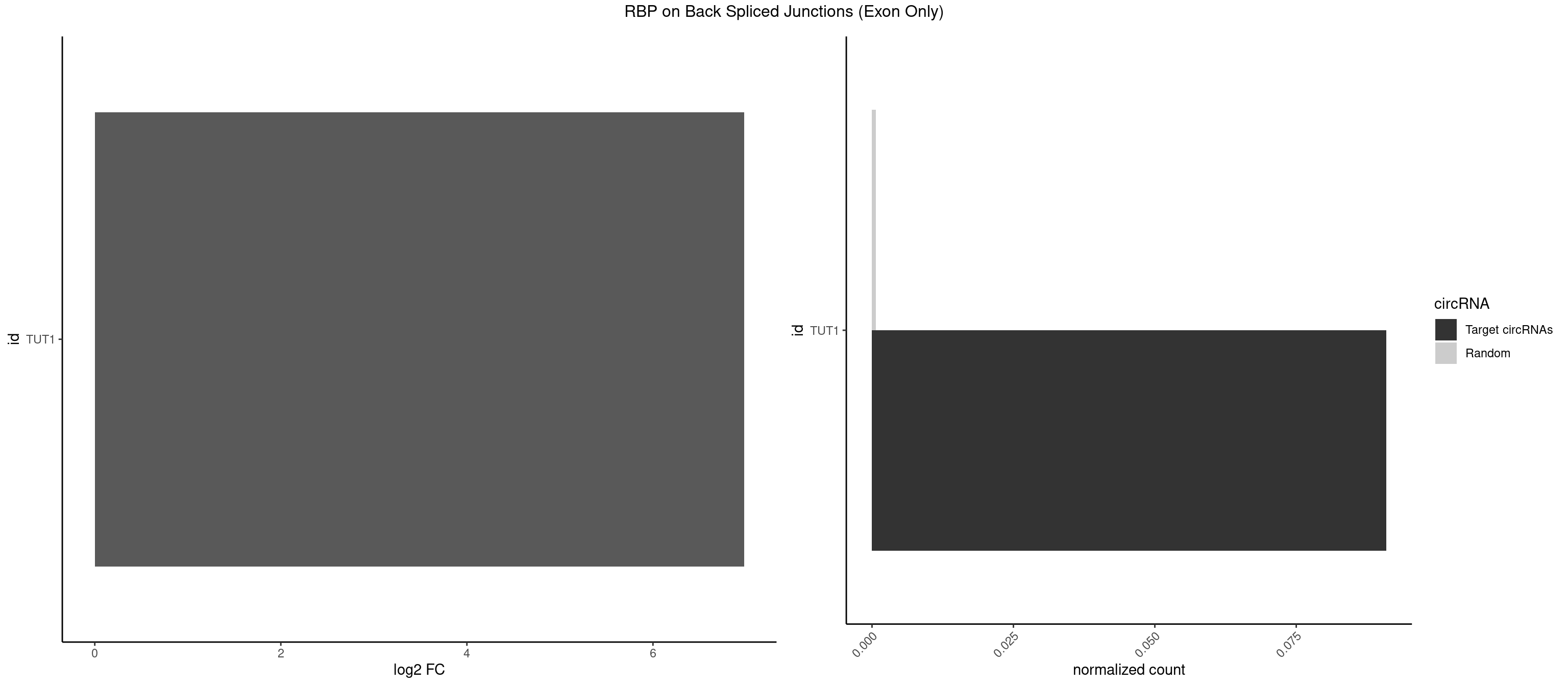
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TUT1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.97936 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
RBP on BSJ (Exon and Intron)
Plot
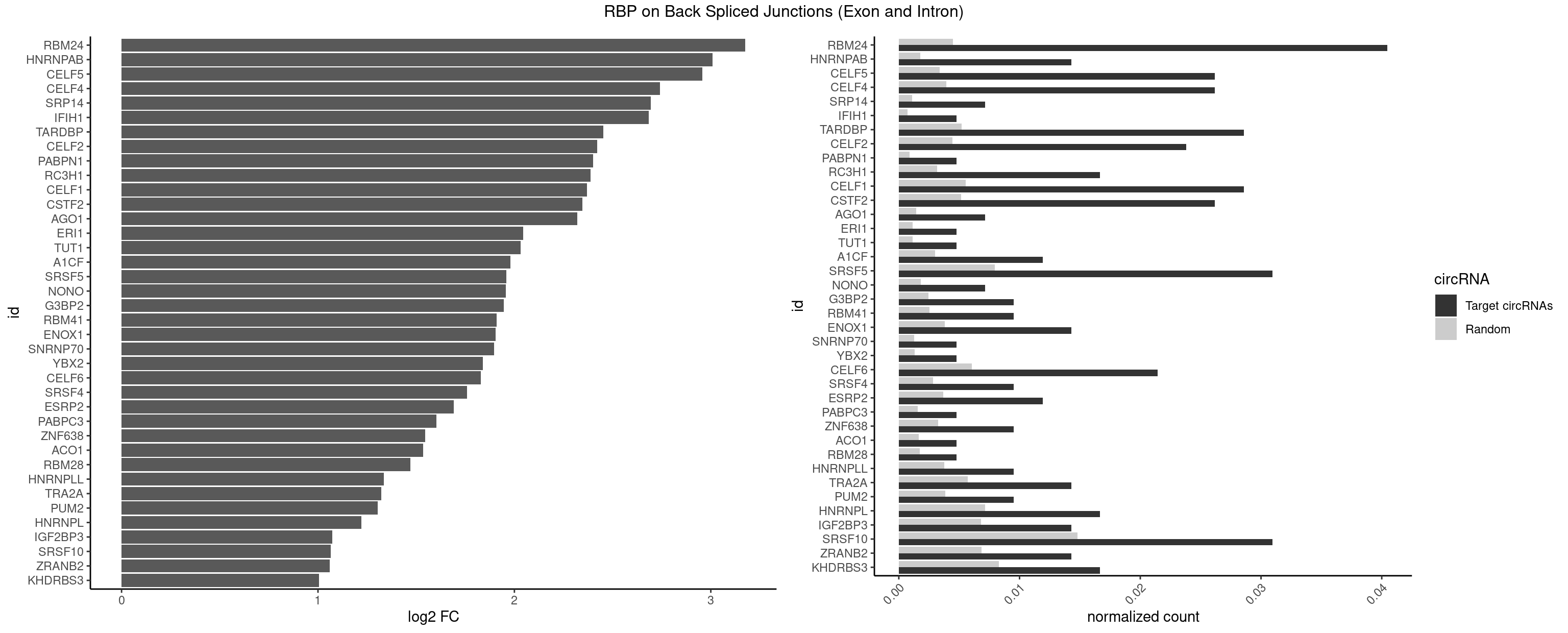
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM24 | 16 | 888 | 0.040476190 | 0.0044790407 | 3.175812 | AGAGUG,AGUGUG,GAGUGG,GAGUGU,GUGUGG,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| HNRNPAB | 5 | 351 | 0.014285714 | 0.0017734784 | 3.009919 | AGACAA,AUAGCA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF5 | 10 | 669 | 0.026190476 | 0.0033756550 | 2.955803 | GUGUGG,GUGUGU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 10 | 776 | 0.026190476 | 0.0039147521 | 2.742049 | GUGUGG,GUGUGU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| TARDBP | 11 | 1034 | 0.028571429 | 0.0052146312 | 2.453936 | GAAUGG,GUGUGU,GUUGUG,GUUUUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF2 | 9 | 881 | 0.023809524 | 0.0044437727 | 2.421682 | GUGUGU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| RC3H1 | 6 | 631 | 0.016666667 | 0.0031841999 | 2.387963 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF1 | 11 | 1097 | 0.028571429 | 0.0055320435 | 2.368689 | GUGUGU,GUUGUG,UGUGUG,UGUGUU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CSTF2 | 10 | 1022 | 0.026190476 | 0.0051541717 | 2.345230 | GUGUGU,GUUUUG,UGUGUG,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUCAGU,GAUCAG,UCAGUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SRSF5 | 12 | 1578 | 0.030952381 | 0.0079554615 | 1.960033 | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,GGAAGA,UACAGC,UACAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | AGAGGA,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ENOX1 | 5 | 756 | 0.014285714 | 0.0038139863 | 1.905202 | AUACAG,CAGACA,CAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF6 | 8 | 1196 | 0.021428571 | 0.0060308343 | 1.829106 | GUGUGG,UGUGGG,UGUGGU,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| SRSF4 | 3 | 558 | 0.009523810 | 0.0028164047 | 1.757684 | AGAAGA,AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ESRP2 | 4 | 731 | 0.011904762 | 0.0036880290 | 1.690617 | GGGAAU,GGGGAG,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUGU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACAUAC,CAUACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AAAGAA,AGAAGA,AGAGGA,AGGAAG,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UAAAUA,UACAUA,UAGAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAAUAA,AACACA,AAUAAA,ACAUAC,CAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AACACA,ACAUAC,CAUACA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SRSF10 | 12 | 2937 | 0.030952381 | 0.0148024990 | 1.064210 | AAAGAA,AGACAA,AGAGAA,AGAGAG,AGAGGA,CAAAGA,GACAAA,GAGAAG,GAGACA,GAGAGC,GAGAGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AGGUAG,AGGUCU,AGGUUU,GAGGUU,GGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAAUAA,AAUAAA,AUAAAA,AUAAAG,UAAAAC,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.