circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000180198_ENSG00000180098:+:1:28529857:28553737
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000180198_ENSG00000180098:+:1:28529857:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28529858 | 28553737 | 2110 | GACAGGAAGAUGUCACCCAAGCGCAUAGCUAAAAGAAGGUCCCCCCCAGCAGAUGCCAUCCCCAAAAGCAAGAAGGUGAAGGUCUCACACAGGUCCCACAGCACAGAACCCGGCUUGGUGCUGACACUAGGCCAGGGCGACGUGGGCCAGCUGGGGCUGGGUGAGAAUGUGAUGGAGAGGAAGAAGCCGGCCCUGGUAUCCAUUCCGGAGGAUGUUGUGCAGGCUGAGGCUGGGGGCAUGCACACCGUGUGUCUAAGCAAAAGUGGCCAGGUCUAUUCCUUCGGCUGCAAUGAUGAGGGUGCCCUGGGAAGGGACACAUCAGUGGAGGGCUCGGAGAUGGUCCCUGGGAAAGUGGAGCUGCAAGAGAAGGUGGUACAGGUGUCAGCAGGAGACAGUCACACAGCAGCCCUCACCGAUGAUGGCCGUGUCUUCCUCUGGGGCUCCUUCCGGGACAAUAACGGUGUGAUUGGACUGUUGGAGCCCAUGAAGAAGAGCAUGGUGCCUGUGCAGGUGCAGCUGGAUGUGCCUGUGGUAAAGGUGGCCUCAGGAAACGACCACUUGGUGAUGCUGACAGCUGAUGGUGACCUCUACACCUUGGGCUGCGGGGAACAGGGCCAGCUAGGCCGUGUGCCUGAGUUAUUUGCCAACCGUGGUGGCCGGCAAGGCCUCGAACGACUCCUGGUCCCCAAGUGUGUGAUGCUGAAAUCCAGGGGAAGCCGGGGCCACGUGAGAUUCCAGGAUGCCUUUUGUGGUGCCUAUUUCACCUUUGCCAUCUCCCAUGAGGGCCACGUGUACGGCUUCGGCCUCUCCAACUACCAUCAGCUUGGAACUCCGGGCACAGAAUCUUGCUUCAUACCCCAGAACCUAACAUCCUUCAAGAAUUCCACCAAGUCCUGGGUGGGCUUCUCUGGUGGCCAGCACCAUACAGUCUGCAUGGAUUCGGAAGGAAAAGCAUACAGCCUGGGCCGGGCUGAGUAUGGGCGGCUGGGCCUUGGAGAGGGUGCUGAGGAGAAGAGCAUACCCACCCUCAUCUCCAGGCUGCCUGCUGUCUCCUCGGUGGCUUGUGGGGCCUCUGUGGGGUAUGCUGUGACCAAGGAUGGUCGUGUUUUCGCCUGGGGCAUGGGCACCAACUACCAGCUGGGCACAGGGCAGGAUGAGGACGCCUGGAGCCCUGUGGAGAUGAUGGGCAAACAGCUGGAGAACCGUGUGGUCUUAUCUGUGUCCAGCGGGGGCCAGCAUACAGUCUUAUUAGUCAAGGACAAAGAACAGAGCUGAUGAAGCCUCUGAGGGCCUGGCUUCUGUCCUGCACAACCUCCCUCACAGAACAGGGAAGCAGUGACAGCUGCAGAUGGCAGCGGGCCUCUCCCCAGCCCUGAGCACUGUGUCAGUUCCUGCCUUUUCUCAUCAGCAGAACAGAAUCCUUUUCCUCUUUUCCUUCCUCCUCUUUGGAAUUUUCCUGGGACCUACAGAAUAAAGGGGGGGAUGGACAGGGGGUUUUCAAAAGGAACAUGGCUCACUCAGAGCUAUAUGGUUAGACGUUUCUCCCCUUUUCCCUACCUUCCAUGGUCCUGGUUGGCCCUGGCUUUGCCUACUAGAAAACCAAAACUUCCCCCCUGGGGUUUUGUGCCCACUCUCUGAGAAGUUGGGGCUCCAUCAAGCCCCAUUCUAGUCAUGUGCCCCUUUCCUGUCCCUAACAGUCCACAGGCAAACAAAUGGUACAGUCAUAAGAGCCAUCUGUCACGGACCCACGCCCAGAGGAACGUGCAGAAAAAAGCAGAGCUACAUGGCUGUGGGCAACUAUAAGCCAAAUAUUUGGCUCAGAACAGGUGUCCAUGGGACAAAAAAGAACGAUCCUCCACUUGACCAAGAAAAAAGUGAUUCUCCCAGAAGCACAAAGCAUACUCUUGCCCCUCAGGUGUUGCUUGUGUACAUCGUACCCAUCCAUUCGGCUUCACCUGCAGCCAACGGCCUGGAAUCGCAAAGAGACACCACUCUGGGCAGAGCAGAGCAGGCUGGAACCCUACAUGGAUGAGAACUUCAUCUCCAGAGCCUUUGCCACCAUGGGGGAGACCGUAAUGAGCGUCAAAAUUAUCCGAAACCGCCUCACUGGGACAGGAAGAUGUCACCCAAGCGCAUAGCUAAAAGAAGGUCCCCCCCAGC | circ |
| ENSG00000180198_ENSG00000180098:+:1:28529857:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28529858 | 28553737 | 22 | CGCCUCACUGGGACAGGAAGAU | bsj |
| ENSG00000180198_ENSG00000180098:+:1:28529857:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28529658 | 28529867 | 210 | GUUAUGAAGGAUACAGUAGAGAAUAUUUGUGUAAUUAAUCUGUGGGUGCAUCCAUUAUUCUGUUCUUGGGAUACAUUUUGAGAAGUGGAAUUGUUGGGCAAUUCCUCUUAACGUAUUUCUAGAGUGUUUGAUAAAUAUUGUCUGAUUGGCCCAGGAAAAUGUUUGCCAUUUCUCAUAUGUAGUAUUUGACUGACUUUCAGGACAGGAAGA | ie_up |
| ENSG00000180198_ENSG00000180098:+:1:28529857:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28553728 | 28553937 | 210 | GCCUCACUGGGUAAGUCUCAUCUCAGGUCUCUCUUAAUACAUCUCGUUGCAGCUGUCAUGUACGAGAAUGUAAACAUCGUAGUCAUGGCUUCUCACUACCCCUAGUAAUAGUCACUGUAAAAUUAUAACAGCUAACAUUUUGAGGUCCCAAUGAGUGGAAGGAUGGCAUGUAGCUUAAGGGUAGGACUUGCCAGGCGUUGUGACUCACAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
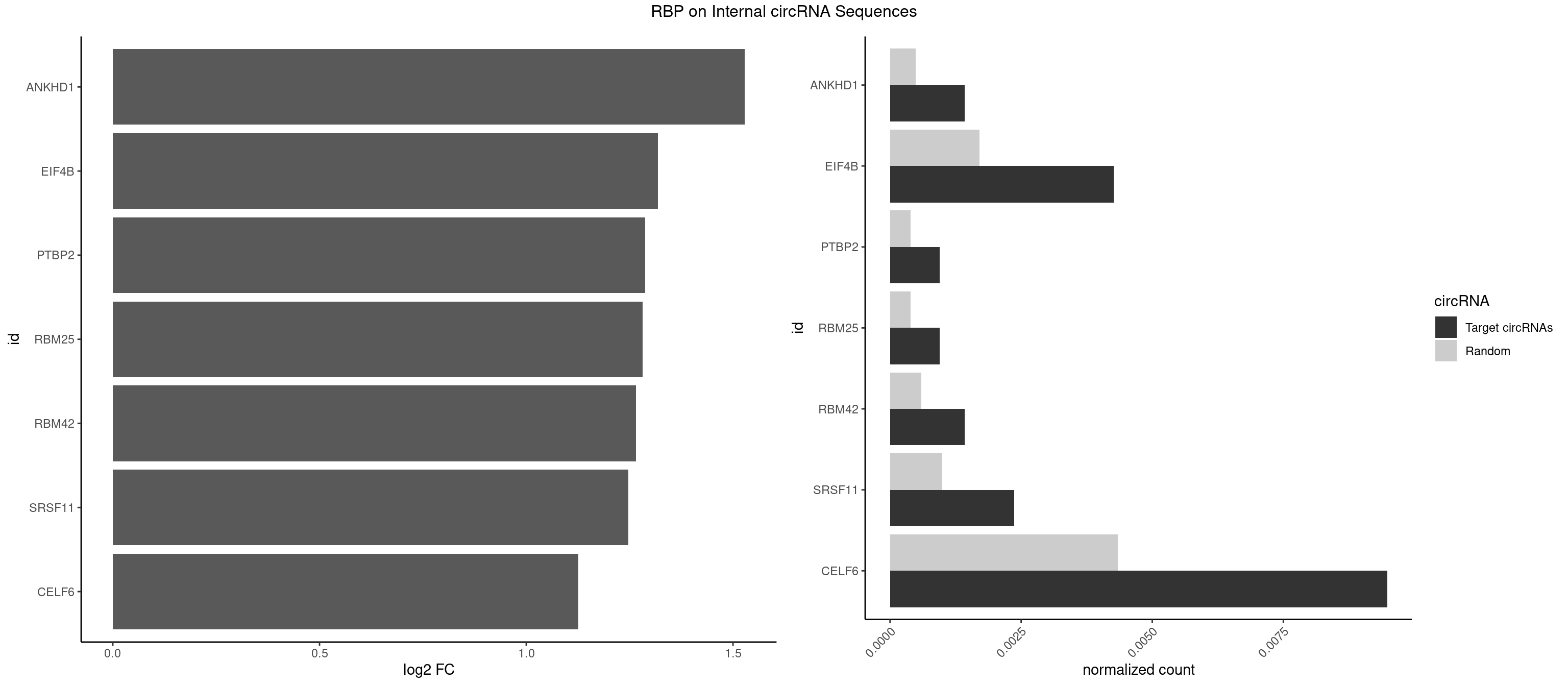
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.0014218009 | 0.0004927293 | 1.528852 | AGACGU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| EIF4B | 8 | 1179 | 0.0042654028 | 0.0017100607 | 1.318634 | CUCGGA,CUUGGA,GUUGGA,UCGGAA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PTBP2 | 1 | 267 | 0.0009478673 | 0.0003883867 | 1.287191 | CUCUCU | CUCUCU |
| RBM25 | 1 | 268 | 0.0009478673 | 0.0003898359 | 1.281818 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBM42 | 2 | 407 | 0.0014218009 | 0.0005912752 | 1.265818 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRSF11 | 4 | 688 | 0.0023696682 | 0.0009985015 | 1.246849 | AAGAAG | AAGAAG |
| CELF6 | 19 | 2997 | 0.0094786730 | 0.0043447134 | 1.125424 | GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAU,UGUGGG,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
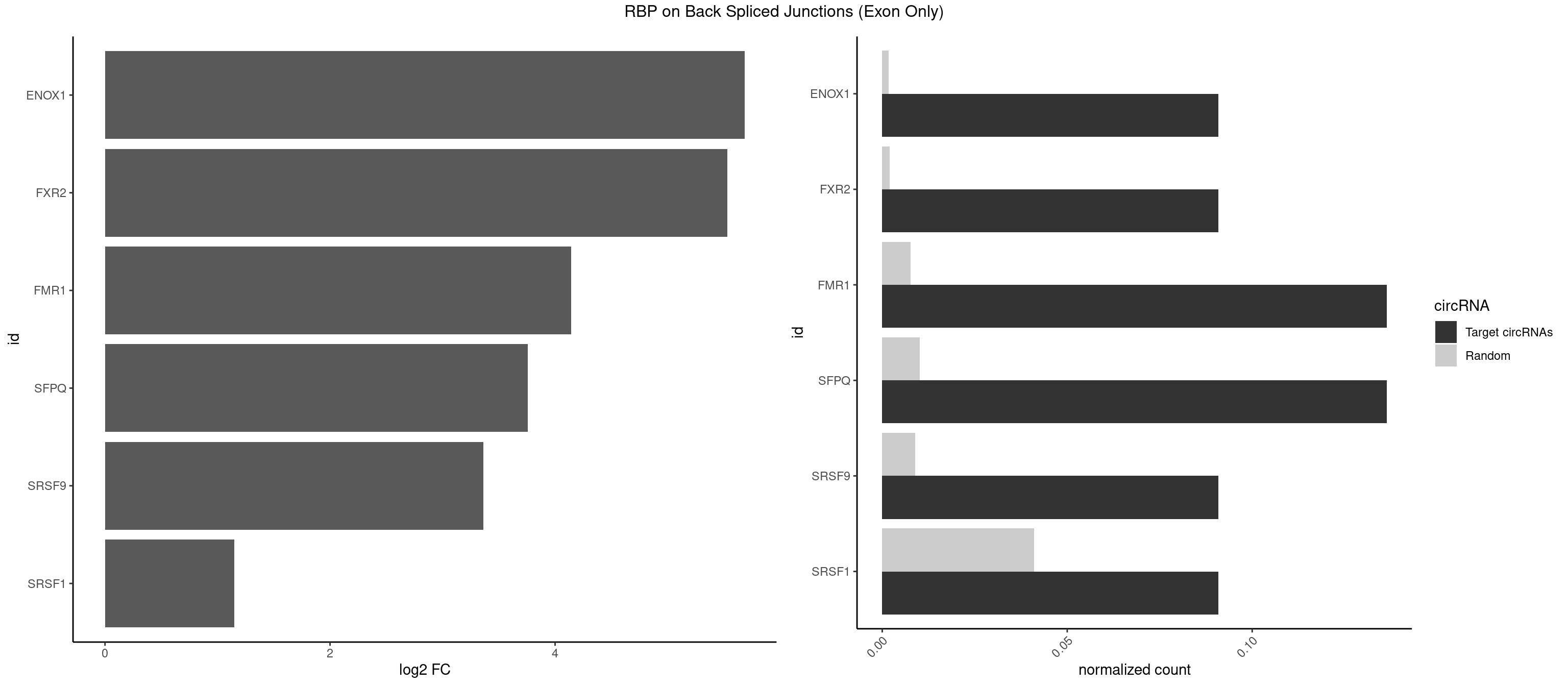
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ENOX1 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | GGACAG | AAGACA,AGACAG,AGGACA,AGUACA,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,UAGACA,UGGACA |
| FXR2 | 1 | 29 | 0.09090909 | 0.001964894 | 5.531901 | GGACAG | AGACAA,AGACAG,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGGA,GGACAA,GGACAG,GGACGA,UGACAA,UGACAG,UGACGA |
| FMR1 | 2 | 117 | 0.13636364 | 0.007728583 | 4.141111 | ACUGGG,GGACAG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SFPQ | 2 | 153 | 0.13636364 | 0.010086455 | 3.756968 | ACUGGG,CUGGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF9 | 1 | 134 | 0.09090909 | 0.008842023 | 3.361976 | GGACAG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SRSF1 | 1 | 625 | 0.09090909 | 0.041000786 | 1.148773 | GGACAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
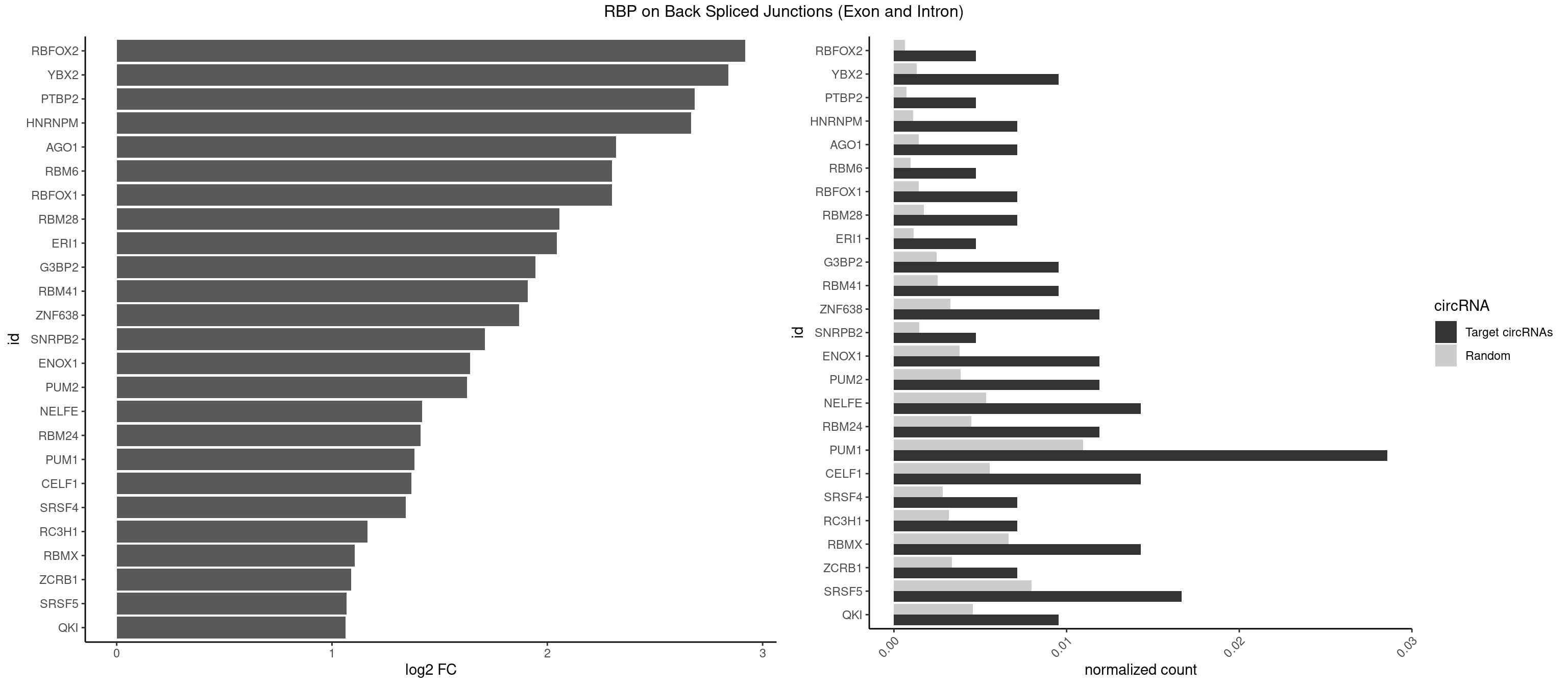
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| YBX2 | 3 | 263 | 0.009523810 | 0.0013301088 | 2.839994 | AACAUC,ACAUCG,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| HNRNPM | 2 | 222 | 0.007142857 | 0.0011235389 | 2.668451 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGU,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUA,AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | CGUUGU,GUUCUU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAUACA,AGGACA,AUACAG,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | UAAAUA,UACAUC,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUCU,GCUAAC,GGUCUC,GUCUCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,GAGUGG,GAGUGU,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUAUU,AAUGUU,AAUUGU,GAAUUG,GUAAUA,UAAAUA,UACAUC,UGUAAA,UGUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | GUUGUG,UGUCUG,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AGGAAG,AGUGUU,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AACAGC,AGGAAG,GGAAGA,UACAUC,UGCAGC,UGCAUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | CUAACA,CUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.