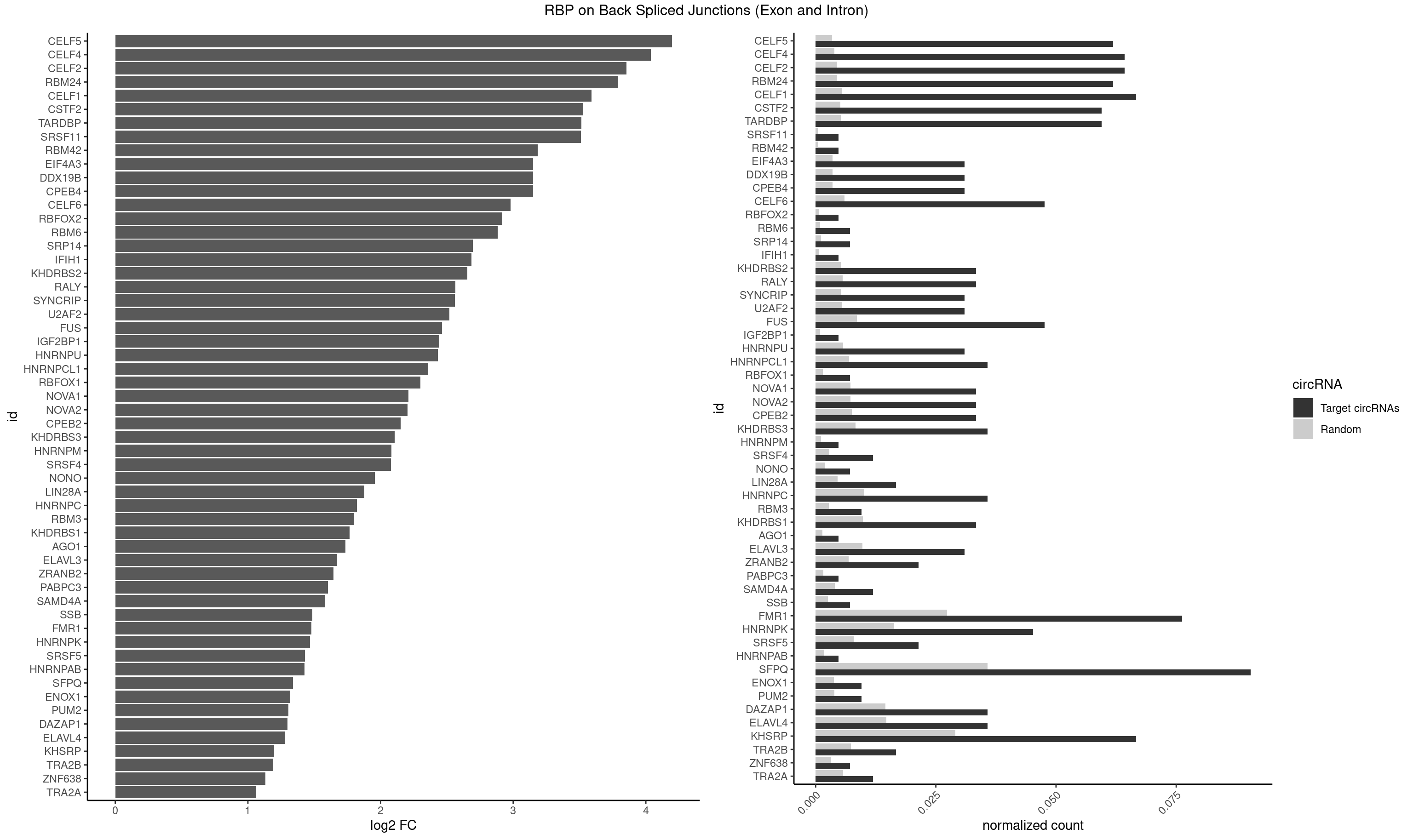
| CELF5 |
25 |
669 |
0.061904762 |
0.0033756550 |
4.196811 |
GUGUGG,GUGUGU,UGUGUG |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 |
26 |
776 |
0.064285714 |
0.0039147521 |
4.037505 |
GGUGUG,GUGUGG,GUGUGU,UGUGUG |
GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF2 |
26 |
881 |
0.064285714 |
0.0044437727 |
3.854641 |
GUCUGU,GUGUGU,UGUGUG,UUGUGU |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 |
25 |
888 |
0.061904762 |
0.0044790407 |
3.788789 |
GUGUGG,GUGUGU,UGUGUG |
AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 |
27 |
1097 |
0.066666667 |
0.0055320435 |
3.591081 |
GUGUGU,GUUUGU,UGUCUG,UGUGUG,UUGUGU |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CSTF2 |
24 |
1022 |
0.059523810 |
0.0051541717 |
3.529654 |
GUGUGU,UGUGUG |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP |
24 |
1034 |
0.059523810 |
0.0052146312 |
3.512830 |
GUGUGU,UGUGUG |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF11 |
1 |
82 |
0.004761905 |
0.0004181782 |
3.509349 |
AAGAAG |
AAGAAG |
| RBM42 |
1 |
103 |
0.004761905 |
0.0005239823 |
3.183949 |
AACUAC |
AACUAA,AACUAC,ACUAAG,ACUACG |
| CPEB4 |
12 |
691 |
0.030952381 |
0.0034864974 |
3.150200 |
UUUUUU |
UUUUUU |
| DDX19B |
12 |
691 |
0.030952381 |
0.0034864974 |
3.150200 |
UUUUUU |
UUUUUU |
| EIF4A3 |
12 |
691 |
0.030952381 |
0.0034864974 |
3.150200 |
UUUUUU |
UUUUUU |
| CELF6 |
19 |
1196 |
0.047619048 |
0.0060308343 |
2.981109 |
GUGGGG,GUGGUG,GUGUGG,UGUGGG,UGUGUG |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBFOX2 |
1 |
124 |
0.004761905 |
0.0006297864 |
2.918604 |
UGCAUG |
UGACUG,UGCAUG |
| RBM6 |
2 |
191 |
0.007142857 |
0.0009673519 |
2.884389 |
AUCCAA,UAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SRP14 |
2 |
218 |
0.007142857 |
0.0011033857 |
2.694564 |
CUGUAG,GCCUGU |
CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 |
1 |
146 |
0.004761905 |
0.0007406288 |
2.684716 |
GGCCCU |
CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| KHDRBS2 |
13 |
1051 |
0.033333333 |
0.0053002821 |
2.652825 |
AAUAAA,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RALY |
13 |
1117 |
0.033333333 |
0.0056328094 |
2.565039 |
UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP |
12 |
1041 |
0.030952381 |
0.0052498992 |
2.559689 |
UUUUUU |
AAAAAA,UUUUUU |
| U2AF2 |
12 |
1071 |
0.030952381 |
0.0054010480 |
2.518739 |
UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| FUS |
19 |
1711 |
0.047619048 |
0.0086255542 |
2.464850 |
CGGUGG,GGGUGA,GGGUGC,GGGUGG,UGGUGG,UGGUGU,UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| IGF2BP1 |
1 |
173 |
0.004761905 |
0.0008766626 |
2.441445 |
AGCACC |
AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPU |
12 |
1136 |
0.030952381 |
0.0057285369 |
2.433812 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPCL1 |
14 |
1381 |
0.035714286 |
0.0069629182 |
2.358737 |
AUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RBFOX1 |
2 |
287 |
0.007142857 |
0.0014510278 |
2.299426 |
AGCAUG,UGCAUG |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| NOVA1 |
13 |
1428 |
0.033333333 |
0.0071997179 |
2.210953 |
AGCACC,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 |
13 |
1434 |
0.033333333 |
0.0072299476 |
2.204908 |
AGCACC,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| CPEB2 |
13 |
1487 |
0.033333333 |
0.0074969770 |
2.152585 |
AUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| KHDRBS3 |
14 |
1646 |
0.035714286 |
0.0082980653 |
2.105654 |
AAUAAA,AUAAAG,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPM |
1 |
222 |
0.004761905 |
0.0011235389 |
2.083489 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| SRSF4 |
4 |
558 |
0.011904762 |
0.0028164047 |
2.079612 |
AAGAAG,AGGAAG,GAAGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO |
2 |
364 |
0.007142857 |
0.0018389762 |
1.957598 |
AGAGGA,GAGAGG |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| LIN28A |
6 |
900 |
0.016666667 |
0.0045395002 |
1.876360 |
CGGAGU,GGAGGG,UGGAGA,UGGAGG,UGGAGU |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPC |
14 |
2006 |
0.035714286 |
0.0101118501 |
1.820454 |
AUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM3 |
3 |
541 |
0.009523810 |
0.0027307537 |
1.802240 |
AAAACU,AAACUA,GAAACG |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| KHDRBS1 |
13 |
1945 |
0.033333333 |
0.0098045143 |
1.765448 |
GAAAAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| AGO1 |
1 |
283 |
0.004761905 |
0.0014308746 |
1.734641 |
GAGGUA |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ELAVL3 |
12 |
1928 |
0.030952381 |
0.0097188634 |
1.671191 |
UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ZRANB2 |
8 |
1360 |
0.021428571 |
0.0068571141 |
1.643862 |
AGGUAA,GGGUGA,GGUGGU,GUGGUG,UAAAGG,UGGUGG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| PABPC3 |
1 |
310 |
0.004761905 |
0.0015669085 |
1.603618 |
GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| SAMD4A |
4 |
790 |
0.011904762 |
0.0039852882 |
1.578783 |
CUGGAC,CUGGCC,GCUGGA |
CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SSB |
2 |
505 |
0.007142857 |
0.0025493753 |
1.486358 |
GCUGUU,UGCUGU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| FMR1 |
31 |
5430 |
0.076190476 |
0.0273629585 |
1.477386 |
AAGGAA,ACUGGG,ACUGGU,AGGAAG,AGGAGG,CUGAGG,CUGGUG,GACUGG,GCUGAG,GCUGGG,GGCUGA,GGCUGG,GGGCUG,GUGGCU,UGGAGU,UGGCUG,UUUUUU |
AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| HNRNPK |
18 |
3244 |
0.045238095 |
0.0163492543 |
1.468313 |
ACCCCA,CAACCC,CCAACC,GCCCAG,GCCCCU,UCCCGA,UUUUUU |
AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| SRSF5 |
8 |
1578 |
0.021428571 |
0.0079554615 |
1.429518 |
AAGAAG,AGGAAG,AUAAAG,GAAGAA,GGAAGA,UAAAGG,UACAGC |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| HNRNPAB |
1 |
351 |
0.004761905 |
0.0017734784 |
1.424957 |
AUAGCA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SFPQ |
37 |
7087 |
0.090476190 |
0.0357114067 |
1.341153 |
AAGGAA,ACUGGG,AGAGGA,CUGGAG,CUGGGA,GAAGAA,GAAGCA,GACUGG,GAGAGG,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGG,GGUAAG,GUAAGA,GUGGUG,UGCAGG,UGGAGA,UGGAGG,UGGUGG,UGGUGU,UUUUUU |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| ENOX1 |
3 |
756 |
0.009523810 |
0.0038139863 |
1.320239 |
GUACAG,UGUACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM2 |
3 |
764 |
0.009523810 |
0.0038542926 |
1.305073 |
GUACAU,UGUACA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| DAZAP1 |
14 |
2878 |
0.035714286 |
0.0145052398 |
1.299927 |
AGGAAG,AGGUAA,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL4 |
14 |
2916 |
0.035714286 |
0.0146966949 |
1.281010 |
UCUAAU,UUUGUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHSRP |
27 |
5764 |
0.066666667 |
0.0290457477 |
1.198639 |
CUGUGU,GUGUGU,UGCAUG,UGUGUG,UUGUGU |
ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| TRA2B |
6 |
1447 |
0.016666667 |
0.0072954454 |
1.191898 |
AAGAAG,AAGAAU,AAGGAA,AGGAAG,GAAGAA,UAAGAA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ZNF638 |
2 |
646 |
0.007142857 |
0.0032597743 |
1.131729 |
GGUUCU,UGUUCU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| TRA2A |
4 |
1133 |
0.011904762 |
0.0057134220 |
1.059112 |
AAGAAG,AGAGGA,AGGAAG,GAAGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |