circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000019995:+:10:124942456:124943307
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000019995:+:10:124942456:124943307 | ENSG00000019995 | MSTRG.4787.1 | + | 10 | 124942457 | 124943307 | 851 | CUUCCUGCCUGACACAGCUCACUUCAAGAAGUGCACAAUGUCAGAACGUGGAAUUAAGUGGGCUUGUGAAUAUUGUACGUAUGAAAACUGGCCAUCUGCAAUCAAGUGUACUAUGUGUCGUGCCCAAAGACCUAGUGGAACAAUUAUUACAGAAGAUCCAUUUAAAAGUGGUUCAAGUGAUGUUGGUAGAGAUUGGGAUCCUUCCAGCACCGAAGGAGGAAGUAGUCCUUUGAUAUGUCCAGACUCUAGUGCAAGACCAAGGGUGAAAUCUUCGUAUAGCAUGGAAAAUGCAAAUAAGUGGUCAUGCCACAUGUGUACAUAUUUGAACUGGCCAAGAGCAAUCAGAUGUACCCAGUGCUUAUCCCAACGUAGGACCAGGAGUCCUACAGAAUCUCCUCAGUCCUCAGGAUCUGGCUCAAGACCAGUUGCUUUUUCUGUUGAUCCUUGUGAGGAAUACAAUGAUAGAAAUAAACUGAACACUAGGACACAGCACUGGACUUGCUCUGUUUGCACAUAUGAAAACUGGGCCAAGGCUAAAAGAUGUGUUGUUUGUGAUCAUCCCAGACCUAAUAACAUUGAAGCAAUAGAAUUGGCAGAGACUGAAGAGGCUUCUUCAAUAAUAAAUGAGCAAGACAGAGCUCGAUGGAGGGGAAGUUGCAGUAGUGGUAAUAGCCAAAGGAGAUCACCUCCUGCUACGAAGCGGGACUCUGAAGUGAAAAUGGAUUUUCAGAGGAUUGAAUUGGCUGGUGCUGUGGGAAGCAAGGAGGAACUUGAAGUAGACUUUAAAAAACUAAAGCAAAUUAAAAACAGGAUGAAAAAGACUGAUUGGCUCUUCCUCAAUGCUUGUGUGGCUUCCUGCCUGACACAGCUCACUUCAAGAAGUGCACAAUGUCAGAACGUG | circ |
| ENSG00000019995:+:10:124942456:124943307 | ENSG00000019995 | MSTRG.4787.1 | + | 10 | 124942457 | 124943307 | 22 | UGCUUGUGUGGCUUCCUGCCUG | bsj |
| ENSG00000019995:+:10:124942456:124943307 | ENSG00000019995 | MSTRG.4787.1 | + | 10 | 124942257 | 124942466 | 210 | CUAAGAUUUUUUCUUUGAGAAUUUAUCUCCAGUGUUUCUAUGGAAAUUAAAAAAGAAAAUUAGGAUAAUUCAAUGUCGAAAUGUUGCAUGCAUCUUUUGAGAAAUUUAUAUUUUGUAGGUUGAAGGACUUGCUUUUUGGGCAGCGUAUUUUUGGAGGUGGAAUGUAGUUAUUUUAAUAACCAUGUCCUAAUUAUUUAUAGCUUCCUGCCU | ie_up |
| ENSG00000019995:+:10:124942456:124943307 | ENSG00000019995 | MSTRG.4787.1 | + | 10 | 124943298 | 124943507 | 210 | GCUUGUGUGGGUAAGUUUCUGUAUUCUGCAUUUUUUGCGUGGGAUGGGAAAAGGUAGUAAAAAUGAUAUGAAAAGAGCUGGGUGCGCUGACACACGUUUGUGGUCUUAGCCACUUGCUUGAAACCAGGAGUUUGAGGCUGUAGUAUGCCAUGAUCGGUCUGUGAAUAGCACUUGCACUCCAGCUUGGGUAACAUAGUGAGACUGUGUCUC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
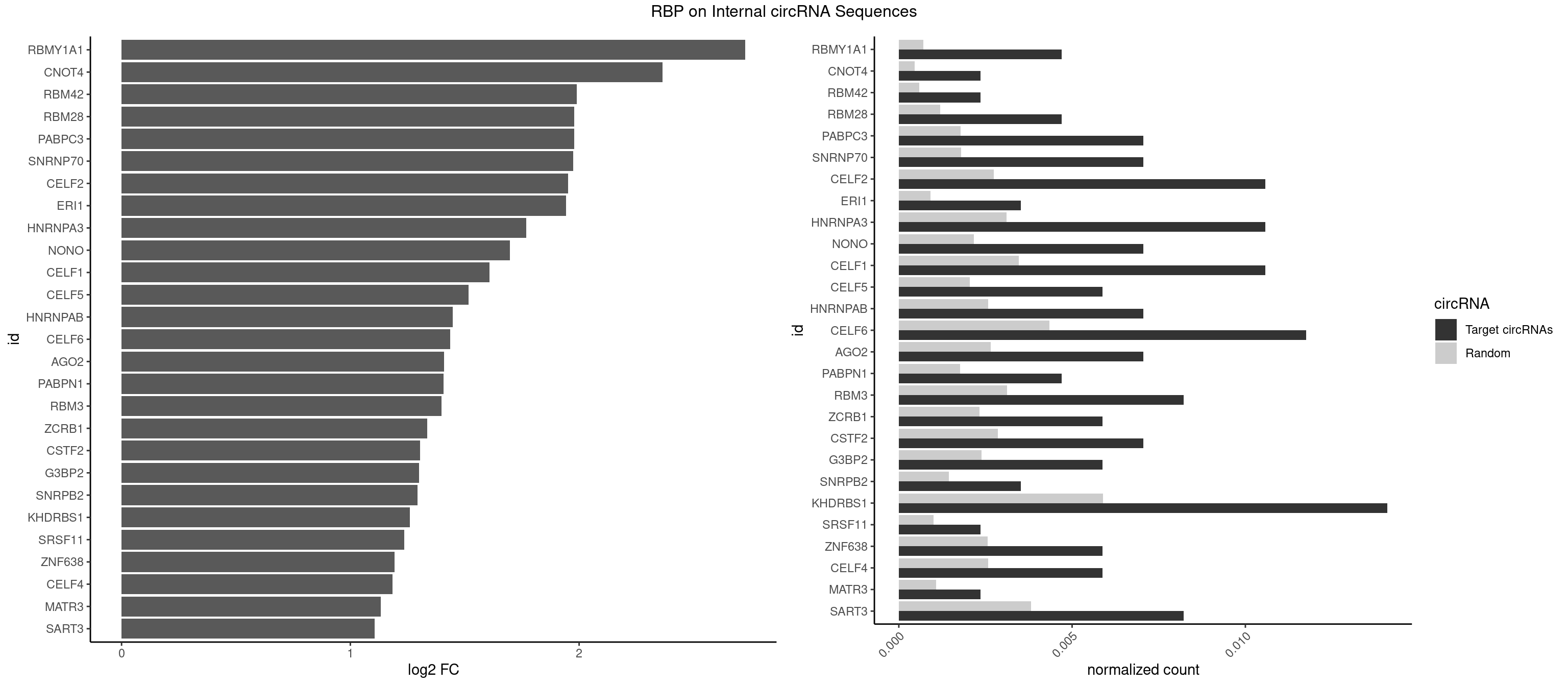
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 3 | 489 | 0.004700353 | 0.0007101099 | 2.726655 | CAAGAC | ACAAGA,CAAGAC |
| CNOT4 | 1 | 314 | 0.002350176 | 0.0004564992 | 2.364085 | GACAGA | GACAGA |
| RBM42 | 1 | 407 | 0.002350176 | 0.0005912752 | 1.990867 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBM28 | 3 | 822 | 0.004700353 | 0.0011926949 | 1.978544 | AGUAGA,AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPC3 | 5 | 1234 | 0.007050529 | 0.0017897669 | 1.977960 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 5 | 1237 | 0.007050529 | 0.0017941145 | 1.974460 | AAUCAA,AUCAAG,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF2 | 8 | 1886 | 0.010575793 | 0.0027346479 | 1.951339 | AUGUGU,GUUGUU,UAUGUG,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ERI1 | 2 | 632 | 0.003525264 | 0.0009173461 | 1.942193 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| HNRNPA3 | 8 | 2140 | 0.010575793 | 0.0031027457 | 1.769149 | AAGGAG,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| NONO | 5 | 1498 | 0.007050529 | 0.0021723567 | 1.698470 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CELF1 | 8 | 2391 | 0.010575793 | 0.0034664959 | 1.609216 | GUGUUG,GUUUGU,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CELF5 | 4 | 1415 | 0.005875441 | 0.0020520728 | 1.517615 | GUGUGG,GUGUUG,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 5 | 1782 | 0.007050529 | 0.0025839306 | 1.448164 | AAAGAC,AAGACA,AUAGCA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF6 | 9 | 2997 | 0.011750881 | 0.0043447134 | 1.435436 | GUGAGG,GUGAUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| AGO2 | 5 | 1830 | 0.007050529 | 0.0026534924 | 1.409839 | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPN1 | 3 | 1222 | 0.004700353 | 0.0017723764 | 1.407084 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM3 | 6 | 2152 | 0.008225617 | 0.0031201361 | 1.398515 | AAAACU,AAACUA,AAGACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ZCRB1 | 4 | 1605 | 0.005875441 | 0.0023274215 | 1.335965 | AAUUAA,AUUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CSTF2 | 5 | 1967 | 0.007050529 | 0.0028520334 | 1.305741 | GUGUUG,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| G3BP2 | 4 | 1644 | 0.005875441 | 0.0023839405 | 1.301349 | AGGAUG,AGGAUU,GGAUGA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRPB2 | 2 | 991 | 0.003525264 | 0.0014376103 | 1.294059 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| KHDRBS1 | 11 | 4064 | 0.014101058 | 0.0058910141 | 1.259215 | AUUUAA,CUAAAA,GAAAAC,UAAAAA,UAAAAG,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SRSF11 | 1 | 688 | 0.002350176 | 0.0009985015 | 1.234932 | AAGAAG | AAGAAG |
| ZNF638 | 4 | 1773 | 0.005875441 | 0.0025708878 | 1.192430 | GUUGGU,GUUGUU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF4 | 4 | 1782 | 0.005875441 | 0.0025839306 | 1.185130 | GUGUGG,GUGUUG,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| MATR3 | 1 | 739 | 0.002350176 | 0.0010724109 | 1.131911 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SART3 | 6 | 2634 | 0.008225617 | 0.0038186524 | 1.107060 | AAAAAA,AAAAAC,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
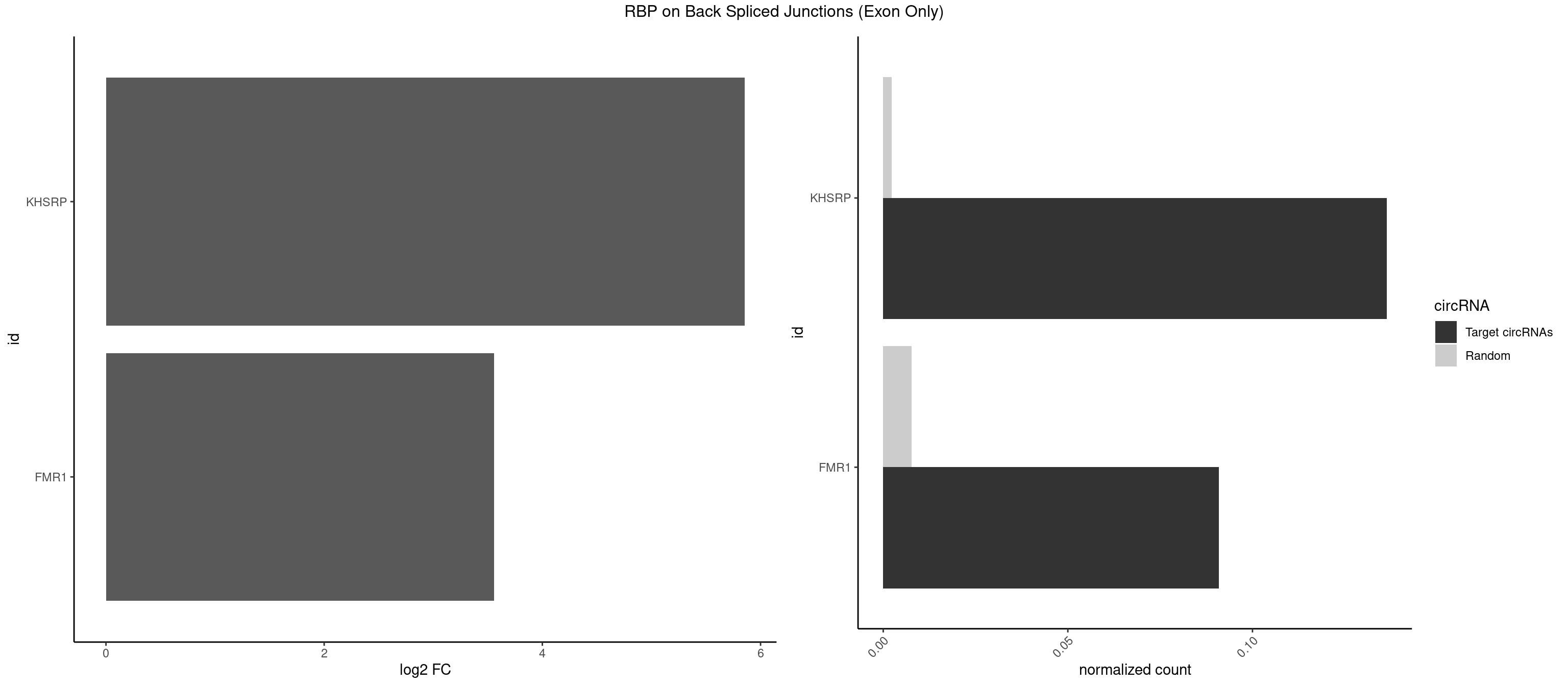
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| KHSRP | 2 | 35 | 0.13636364 | 0.002357873 | 5.853829 | GCUUCC,GGCUUC | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | GUGGCU | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
RBP on BSJ (Exon and Intron)
Plot
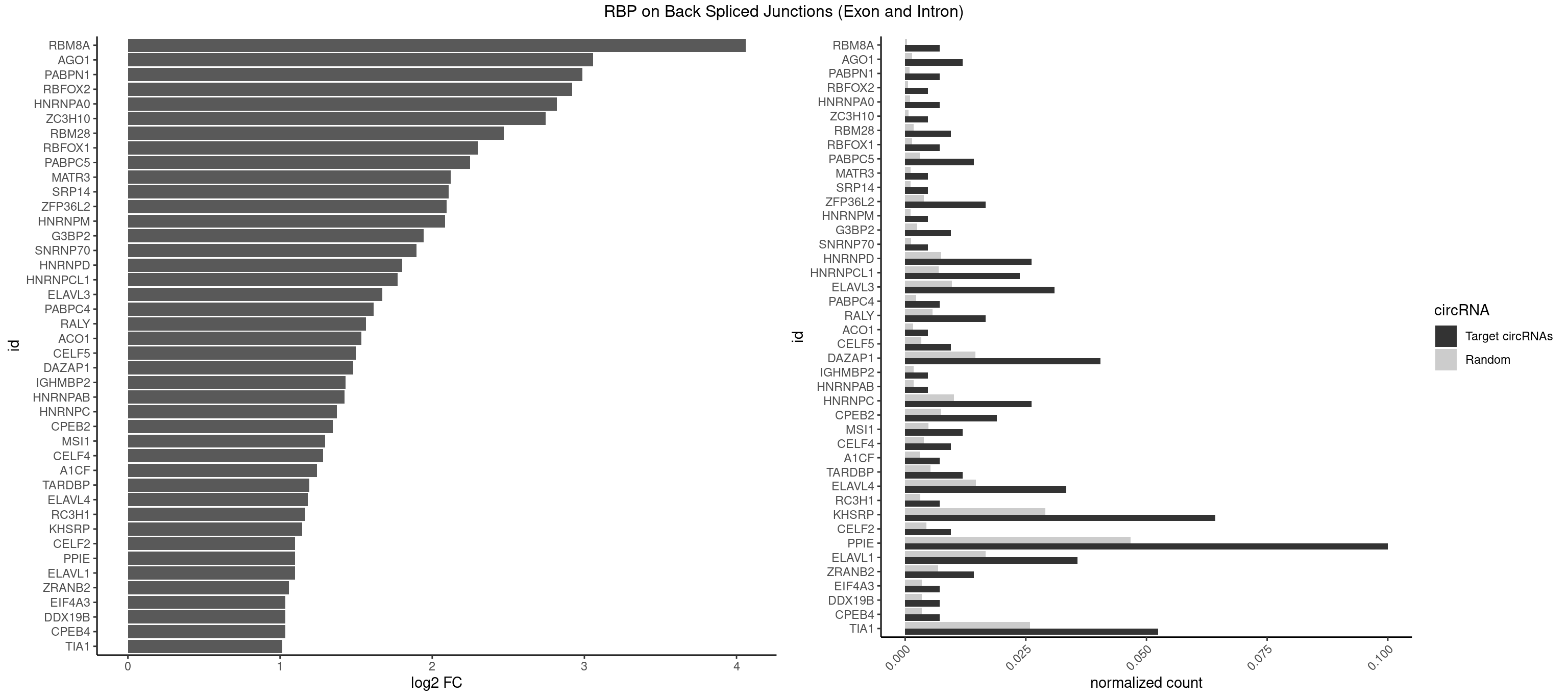
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 2 | 84 | 0.007142857 | 0.0004282547 | 4.059960 | GUGCGC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| AGO1 | 4 | 283 | 0.011904762 | 0.0014308746 | 3.056570 | AGGUAG,GGUAGU,GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM28 | 3 | 340 | 0.009523810 | 0.0017180572 | 2.470761 | UGUAGG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGC,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC5 | 5 | 596 | 0.014285714 | 0.0030078597 | 2.247764 | AGAAAA,AGAAAU,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ZFP36L2 | 6 | 774 | 0.016666667 | 0.0039046755 | 2.093691 | AUUUAU,UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUA,GGAUAA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPD | 10 | 1488 | 0.026190476 | 0.0075020153 | 1.803692 | AAAAAA,AAUUUA,AUUUAU,UAUUUA,UUAGGA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPCL1 | 9 | 1381 | 0.023809524 | 0.0069629182 | 1.773775 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| ELAVL3 | 12 | 1928 | 0.030952381 | 0.0097188634 | 1.671191 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGG,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| DAZAP1 | 16 | 2878 | 0.040476190 | 0.0145052398 | 1.480499 | AAAAAA,AAUUUA,AGGAUA,AGGUAG,AGGUUG,AGUAAA,AGUAUG,AGUUUG,UAGGAU,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,CUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUAG,AGGUGG,UAGUAA,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGG,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGU,GUGAAU,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ELAVL4 | 13 | 2916 | 0.033333333 | 0.0146966949 | 1.181474 | AAAAAA,AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUGUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| KHSRP | 26 | 5764 | 0.064285714 | 0.0290457477 | 1.146171 | AGCUUC,AUAUUU,AUUAUU,AUUUAU,AUUUUA,CAGCUU,CUGCCU,CUGUAU,CUGUGU,GCUUCC,UAGGUU,UAGUAU,UAUAUU,UAUUUA,UAUUUU,UGCAUG,UGUGUG,UUAUUU,UUGUGU,UUUAUA | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUCUGU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PPIE | 41 | 9262 | 0.100000000 | 0.0466696896 | 1.099442 | AAAAAA,AAAAAU,AAAAUU,AAAUUA,AAAUUU,AAUUAA,AAUUAU,AAUUUA,AUAAUU,AUAUUU,AUUAAA,AUUAUU,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAUAA,UAAUUA,UAUAUU,UAUUUA,UAUUUU,UUAAAA,UUAAUA,UUAUAU,UUAUUU,UUUAAU,UUUAUA,UUUUAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AAAGGU,AGGUAG,CGGUCU,GGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUG,AUUUUU,CUUUUG,CUUUUU,UAUUUU,UUAUUU,UUUUCU,UUUUGG,UUUUGU,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.