circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000198879:-:10:7276891:7285954
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000198879:-:10:7276891:7285954 | ENSG00000198879 | ENST00000397167 | - | 10 | 7276892 | 7285954 | 434 | CAAUCAAAGAGAAGUACACAGACUGGACAGAAUUUCUCAUACGUGACUUGACUGGUUCGAGGACAGCACCCGCCAACCUCCUGGAAGGUCCUCUGCGAGGGAAAGGCCCUAUAGACCUCAUUACAGUUGGUUCCUUAAUAGAACUUCAGGAUUCCCAGAACCCUUUUCAGUACUGGAUAGUUAGUGUGAUUGAAAAUGUUGGAGGAAGAUUACGCCUUCGCUAUGUGGGAUUGGAGGACACUGAAUCCUAUGACCAGUGGUUGUUUUACUUGGAUUACAGACUUCGACCAGUUGGUUGGUGUCAAGAGAAUAAAUACAGAAUGGACCCACCUUCAGAAAUCUAUCCUUUGAAGAUGGCCUCUGAAUGGAAAUGUACUCUGGAAAAAUCCCUUAUUGAUGCUGCCAAAUUUCCUCUUCCAAUGGAAGUGUUUAAGCAAUCAAAGAGAAGUACACAGACUGGACAGAAUUUCUCAUACGUGACUUG | circ |
| ENSG00000198879:-:10:7276891:7285954 | ENSG00000198879 | ENST00000397167 | - | 10 | 7276892 | 7285954 | 22 | AAGUGUUUAAGCAAUCAAAGAG | bsj |
| ENSG00000198879:-:10:7276891:7285954 | ENSG00000198879 | ENST00000397167 | - | 10 | 7285945 | 7286154 | 210 | GUCAAGUUGCUACUUAGAGACACAUUUUCAGUCUCAGUUUUAGUAUUUACUUAAGCAUGGUGAUGAAAGUUAAGAAAAACAUUGAAAUCAAACCUACAUGGAGAAAACUGCUGUGAUUUUAAGUUGAUUGGAUACUUUGCUGCUGUGUUGAAGUGUUUUUUUUGUUUUGUUUUGUUUUUCUUUCUCCUUUGUCUUGCCAGCAAUCAAAGA | ie_up |
| ENSG00000198879:-:10:7276891:7285954 | ENSG00000198879 | ENST00000397167 | - | 10 | 7276692 | 7276901 | 210 | AGUGUUUAAGGUAAGAUGUUGAUUCUUAGCAAUGUAUCUACCCUUUUUGAGAAUAAAAUCAUCUACAAAGUUUUCCGGCCGGGCGCAGUGGCUCAUGCCUGUAAUCCCAGCACUUUGAGAGGCUGAGGUGGGCGGAUCACGACAUCAGGAGAUCGAGACCAUCCUGGCUAACAUGGUGAAACCCUAUUUCUACUAAAAAUACAAAAAUUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
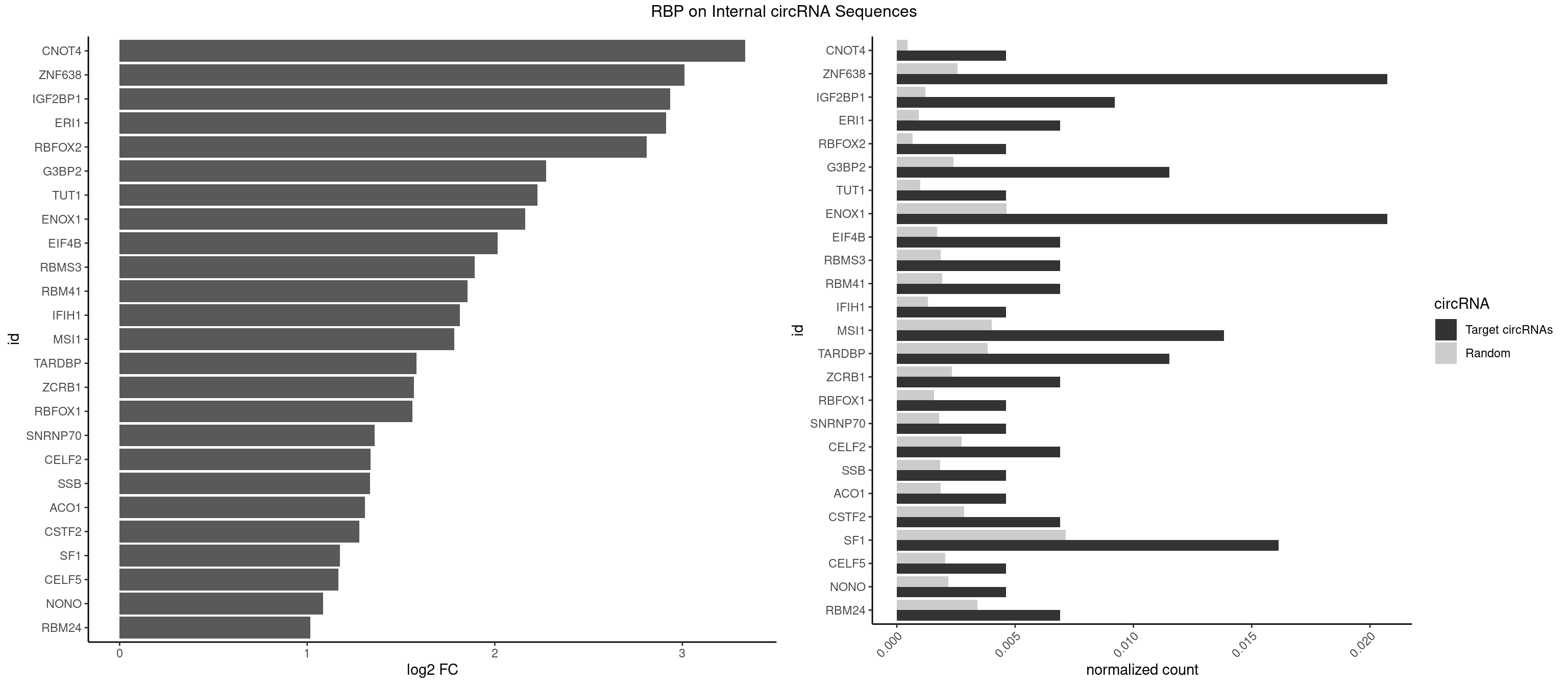
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.004608295 | 0.0004564992 | 3.335549 | GACAGA | GACAGA |
| ZNF638 | 8 | 1773 | 0.020737327 | 0.0025708878 | 3.011891 | GGUUCG,GGUUGG,GGUUGU,GUUGGU,GUUGUU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| IGF2BP1 | 3 | 831 | 0.009216590 | 0.0012057377 | 2.934317 | AGCACC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ERI1 | 2 | 632 | 0.006912442 | 0.0009173461 | 2.913658 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBFOX2 | 1 | 452 | 0.004608295 | 0.0006564894 | 2.811389 | UGACUG | UGACUG,UGCAUG |
| G3BP2 | 4 | 1644 | 0.011520737 | 0.0023839405 | 2.272813 | AGGAUU,GGAUAG,GGAUUA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TUT1 | 1 | 678 | 0.004608295 | 0.0009840095 | 2.227489 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ENOX1 | 8 | 3195 | 0.020737327 | 0.0046316558 | 2.162630 | AAUACA,AGGACA,AGUACA,AUACAG,GGACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| EIF4B | 2 | 1179 | 0.006912442 | 0.0017100607 | 2.015148 | CUUGGA,GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBMS3 | 2 | 1283 | 0.006912442 | 0.0018607779 | 1.893290 | CUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM41 | 2 | 1318 | 0.006912442 | 0.0019115000 | 1.854490 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| IFIH1 | 1 | 904 | 0.004608295 | 0.0013115296 | 1.812983 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| MSI1 | 5 | 2770 | 0.013824885 | 0.0040157442 | 1.783528 | AGGAAG,AGUUAG,AGUUGG,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| TARDBP | 4 | 2654 | 0.011520737 | 0.0038476365 | 1.582189 | GAAUGG,GUUGUU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZCRB1 | 2 | 1605 | 0.006912442 | 0.0023274215 | 1.570463 | GGAUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBFOX1 | 1 | 1077 | 0.004608295 | 0.0015622419 | 1.560615 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRNP70 | 1 | 1237 | 0.004608295 | 0.0017941145 | 1.360961 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF2 | 2 | 1886 | 0.006912442 | 0.0027346479 | 1.337840 | GUUGUU,UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SSB | 1 | 1260 | 0.004608295 | 0.0018274462 | 1.334404 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ACO1 | 1 | 1283 | 0.004608295 | 0.0018607779 | 1.308327 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CSTF2 | 2 | 1967 | 0.006912442 | 0.0028520334 | 1.277205 | GUGUUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SF1 | 6 | 4931 | 0.016129032 | 0.0071474739 | 1.174155 | ACAGAC,CACAGA,CACUGA,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF5 | 1 | 1415 | 0.004608295 | 0.0020520728 | 1.167151 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| NONO | 1 | 1498 | 0.004608295 | 0.0021723567 | 1.084972 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM24 | 2 | 2357 | 0.006912442 | 0.0034172229 | 1.016371 | AGUGUG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
RBP on BSJ (Exon Only)
Plot
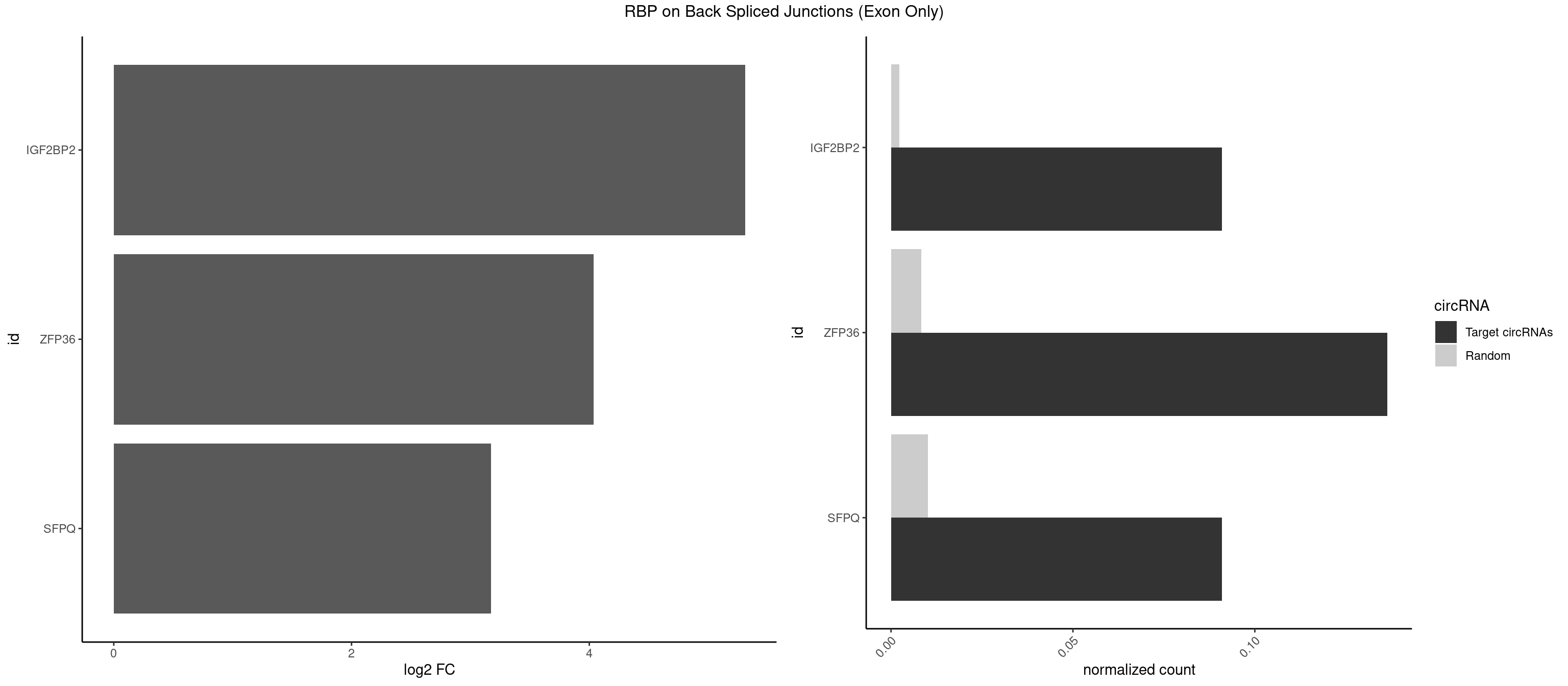
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGF2BP2 | 1 | 34 | 0.09090909 | 0.002292376 | 5.309509 | GCAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
| ZFP36 | 2 | 126 | 0.13636364 | 0.008318051 | 4.035070 | AAGCAA,UAAGCA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | AAGCAA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
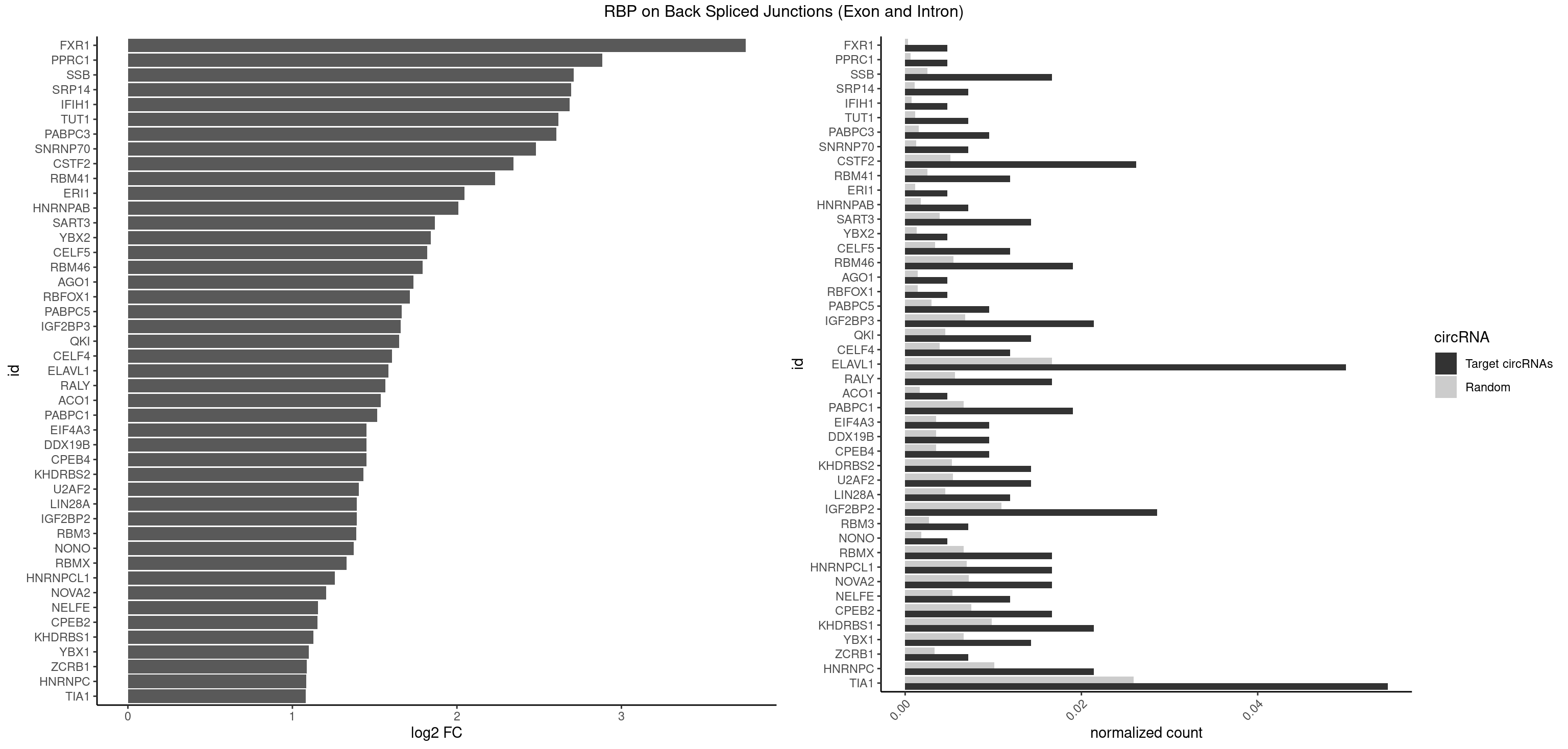
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| PPRC1 | 1 | 127 | 0.004761905 | 0.0006449012 | 2.884389 | GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| SSB | 6 | 505 | 0.016666667 | 0.0025493753 | 2.708750 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CSTF2 | 10 | 1022 | 0.026190476 | 0.0051541717 | 2.345230 | GUGUUG,GUGUUU,GUUUUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACAUG,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SART3 | 5 | 777 | 0.014285714 | 0.0039197904 | 1.865725 | AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUUG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM46 | 7 | 1091 | 0.019047619 | 0.0055018138 | 1.791631 | AAUCAA,AAUCAU,AUCAAA,AUGAAA,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 8 | 1348 | 0.021428571 | 0.0067966546 | 1.656639 | AAAAAC,AAAACA,AAAAUC,AAAUAC,AAAUCA,AAUACA,CAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AAUCAU,AUCUAC,CUAACA,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUUG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL1 | 20 | 3309 | 0.050000000 | 0.0166767432 | 1.584091 | UAUUUA,UGAUUU,UGUUUU,UUGAUU,UUGUUU,UUUAGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PABPC1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | AAAAAC,AGAAAA,CAAACC,CUAACA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| KHDRBS2 | 5 | 1051 | 0.014285714 | 0.0053002821 | 1.430432 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.0054010480 | 1.403262 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGA,GGAGAA,GGAGAU,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| IGF2BP2 | 11 | 2163 | 0.028571429 | 0.0109028617 | 1.389866 | AAAAAC,AAAACA,AAAAUC,AAAUAC,AAAUCA,AAUACA,CAAUCA,GAAAAC,GAAAUC,GCAAUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AAGUGU,AGUGUU,AUCAAA,AUCCCA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AUCAUC,GAGACA,GAUCAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUGGCU,GCUAAC,GGCUAA,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| CPEB2 | 6 | 1487 | 0.016666667 | 0.0074969770 | 1.152585 | CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUAAAA,AUUUAC,CUAAAA,GAAAAC,UAAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACAUCA,AUCAUC,CAGCAA,CCAGCA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | ACUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.0101118501 | 1.083489 | CUUUUU,GGAUAC,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 | 22 | 5140 | 0.054761905 | 0.0259018541 | 1.080117 | AUUUUC,CUCCUU,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UCCUUU,UCUCCU,UUCUCC,UUUUCU,UUUUGU,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.