circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000075151:-:1:20849414:20855071
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000075151:-:1:20849414:20855071 | ENSG00000075151 | ENST00000684485 | - | 1 | 20849415 | 20855071 | 549 | CCUACAAUUGAUGAAAAAAUUCAGCUGGUACCUAAAGCACAGCUAGGCAGCUGGGGAAAAGGCAGCAGUGGUGGAGCAAAGGCAAGUGAGACUGAUGCCUUACGGUCAAGUGCUUCCAGUUUAAACAGAUUCUCUGCCCUGCAACCUCCAGCACCCUCAGGGUCCACGCCAUCCACGCCUGUAGAGUUUGAUUCCCGAAGGACCUUAACUAGUCGUGGAAGUAUGGGCAGGGAGAAGAAUGACAAGCCCCUUCCAUCUGCAACAGCUCGGCCAAAUACUUUCAUGAGGGGUGGCAGCAGUAAAGACCUGCUAGACAAUCAGUCUCAAGAAGAGCAGCGGAGAGAGAUGCUGGAGACCGUGAAGCAGCUCACAGGAGGUGUGGAUGUGGAGAGGAACAGCACUGAGGCUGAGCGAAAUAAAACAAGGGAGUCAGCAAAACCAGAAAUUUCAGCAAUGUCAGCUCAUGACAAGGCUGCAUUAUCAGAAGAGGAACUGGAGAGGAAGUCGAAAUCUAUCAUUGAUGAAUUUCUACACAUUAAUGAUUUUAAGCCUACAAUUGAUGAAAAAAUUCAGCUGGUACCUAAAGCACAGCUAGGCAG | circ |
| ENSG00000075151:-:1:20849414:20855071 | ENSG00000075151 | ENST00000684485 | - | 1 | 20849415 | 20855071 | 22 | AUGAUUUUAAGCCUACAAUUGA | bsj |
| ENSG00000075151:-:1:20849414:20855071 | ENSG00000075151 | ENST00000684485 | - | 1 | 20855062 | 20855271 | 210 | AGUCCAGGGAAGUGCUUCAGGGGCCACAUGGAUCCCAGUUUAAUGAGAACAGCUCCAAUUUCUGUUUCUAUUAAAUUGGCAUUCCAAACAAAUUUUUGUUGCUAAAAUUGGUUCUACUUCACUGAAGAAGAAAAAAAUAAAAGAAAAACUAUUCUUCUAGAAUAUUUCCUAUAAUUGACUUGUGGUCCCUUAUGUUACAGCCUACAAUUG | ie_up |
| ENSG00000075151:-:1:20849414:20855071 | ENSG00000075151 | ENST00000684485 | - | 1 | 20849215 | 20849424 | 210 | UGAUUUUAAGGUAAAUGAGAUUAAAAAAAAAACACACACACAGUAAGUCCUUUGAUAGUAUAGAAUAACUUGGUUCUGGUCUAAAUCAGGUGUCAUUGUGAACUGAUAAGACUUAGGUUGAGUAAUACAAGUCCUACACAAUUCAGUUUGCUUAACGCUGAUGCAUUUCUGCCAAGUACUCUCUUUUAAUUUGAUACUAUAAUUUACAAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
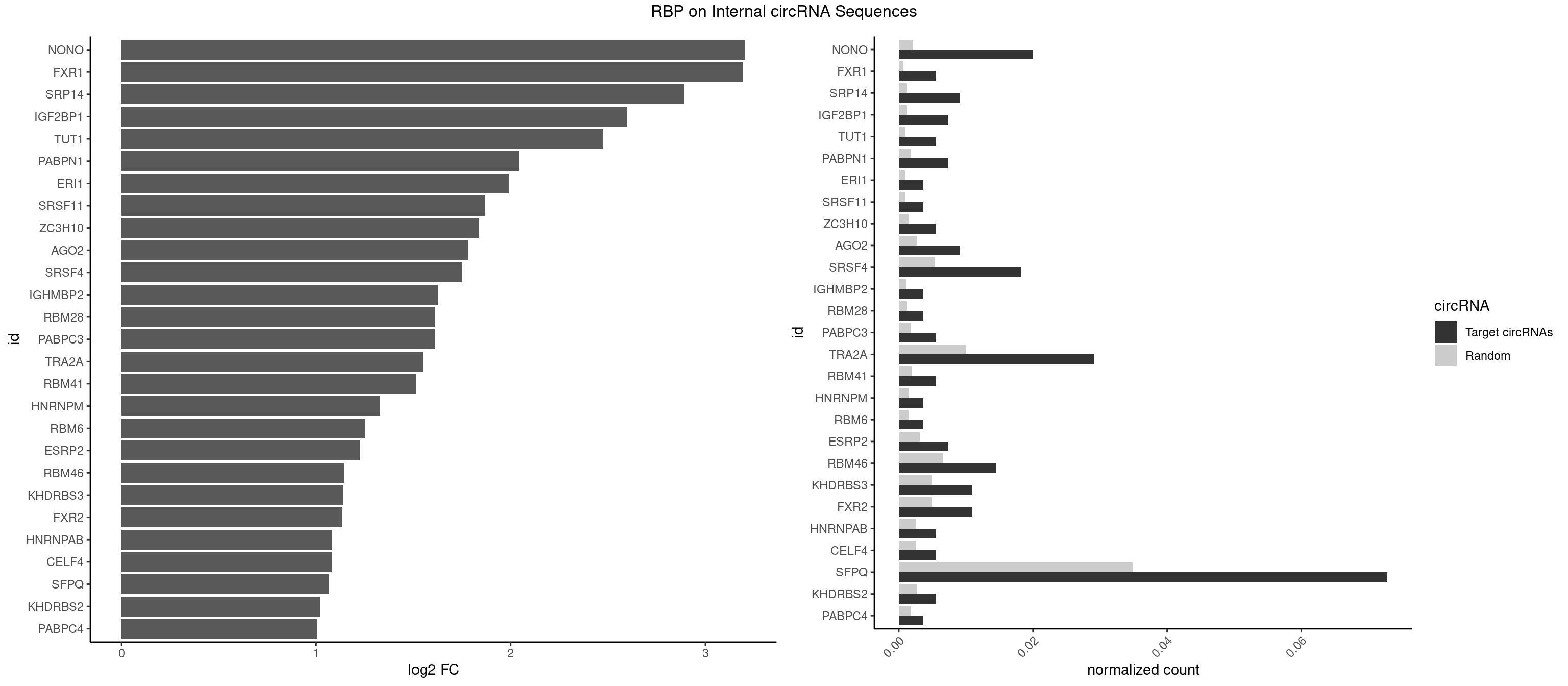
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NONO | 10 | 1498 | 0.020036430 | 0.0021723567 | 3.205293 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| FXR1 | 2 | 411 | 0.005464481 | 0.0005970720 | 3.194108 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRP14 | 4 | 847 | 0.009107468 | 0.0012289250 | 2.889653 | CCUGUA,CGCCUG,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IGF2BP1 | 3 | 831 | 0.007285974 | 0.0012057377 | 2.595206 | AAGCAC,AGCACC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| TUT1 | 2 | 678 | 0.005464481 | 0.0009840095 | 2.473340 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPN1 | 3 | 1222 | 0.007285974 | 0.0017723764 | 2.039437 | AGAAGA | AAAAGA,AGAAGA |
| ERI1 | 1 | 632 | 0.003642987 | 0.0009173461 | 1.989584 | UUUCAG | UUCAGA,UUUCAG |
| SRSF11 | 1 | 688 | 0.003642987 | 0.0009985015 | 1.867285 | AAGAAG | AAGAAG |
| ZC3H10 | 2 | 1053 | 0.005464481 | 0.0015274610 | 1.838949 | GAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| AGO2 | 4 | 1830 | 0.009107468 | 0.0026534924 | 1.779158 | AAAAAA,AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF4 | 9 | 3740 | 0.018214936 | 0.0054214720 | 1.748365 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| IGHMBP2 | 1 | 813 | 0.003642987 | 0.0011796520 | 1.626761 | AAAAAA | AAAAAA |
| RBM28 | 1 | 822 | 0.003642987 | 0.0011926949 | 1.610897 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPC3 | 2 | 1234 | 0.005464481 | 0.0017897669 | 1.610313 | AAAACA,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| TRA2A | 15 | 6871 | 0.029143898 | 0.0099589296 | 1.549131 | AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM41 | 2 | 1318 | 0.005464481 | 0.0019115000 | 1.515379 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 1 | 999 | 0.003642987 | 0.0014492040 | 1.329861 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM6 | 1 | 1054 | 0.003642987 | 0.0015289102 | 1.252618 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ESRP2 | 3 | 2150 | 0.007285974 | 0.0031172377 | 1.224854 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM46 | 7 | 4554 | 0.014571949 | 0.0066011240 | 1.142410 | AAUGAU,AUCAUU,AUGAAA,AUGAAU,AUGAUU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| KHDRBS3 | 5 | 3429 | 0.010928962 | 0.0049707696 | 1.136615 | AAAUAA,AAUAAA,AUAAAA,UAAAAC,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FXR2 | 5 | 3434 | 0.010928962 | 0.0049780156 | 1.134514 | AGACAA,GACAAG,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CELF4 | 2 | 1782 | 0.005464481 | 0.0025839306 | 1.080517 | GGUGUG,GUGUGG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPAB | 2 | 1782 | 0.005464481 | 0.0025839306 | 1.080517 | AAAGAC,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SFPQ | 39 | 24050 | 0.072859745 | 0.0348548043 | 1.063764 | AAGAGC,AAGAGG,AAGGAC,AAGGGA,AGAGAG,AGAGGA,AGGAAC,AGGACC,CUGGAG,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGAGG,GAGGAA,GGAGAG,GUGGUG,UGAAGC,UGAUUU,UGGAAG,UGGAGA,UGGAGC,UGGUGG,UUAAUG,UUGAUG,UUGAUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| KHDRBS2 | 2 | 1858 | 0.005464481 | 0.0026940701 | 1.020297 | AAUAAA,AUAAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PABPC4 | 1 | 1251 | 0.003642987 | 0.0018144033 | 1.005627 | AAAAAA | AAAAAA,AAAAAG |
RBP on BSJ (Exon Only)
Plot
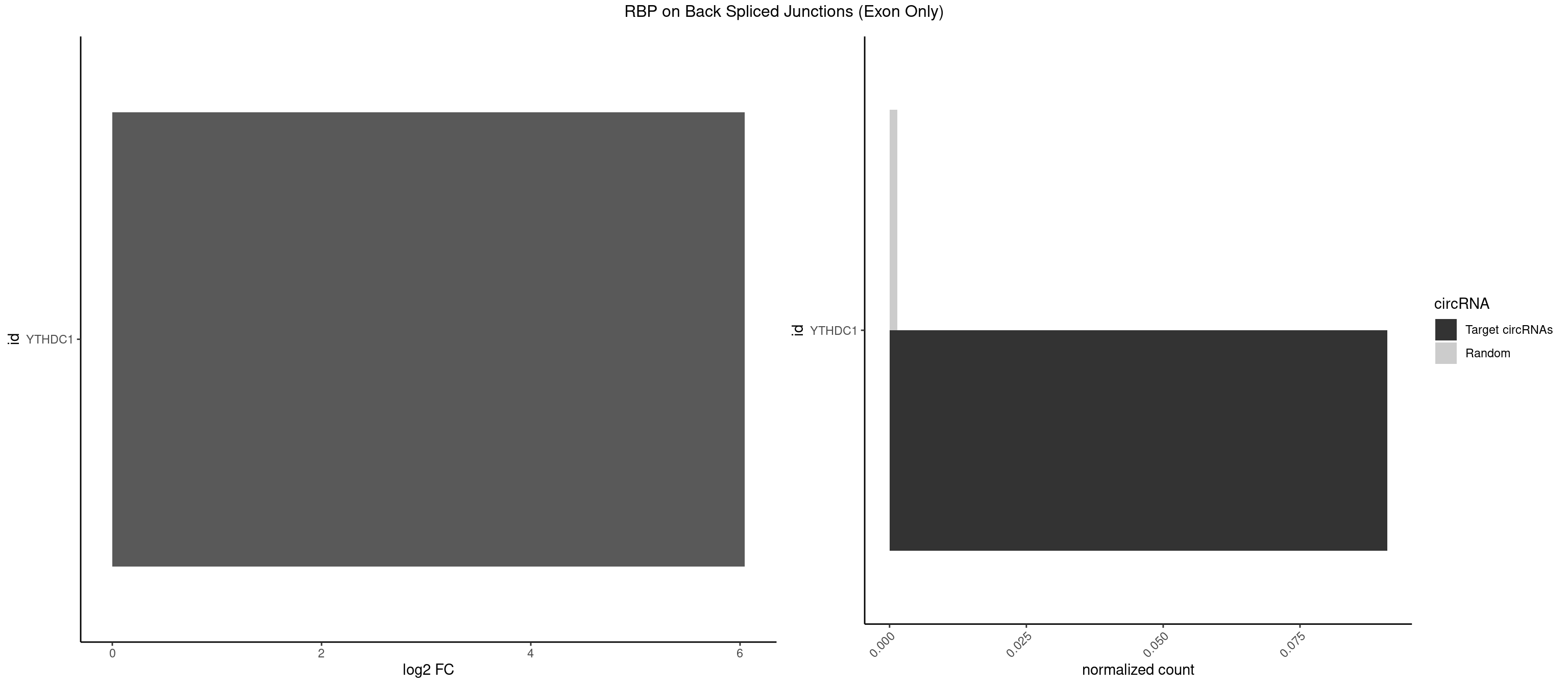
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YTHDC1 | 1 | 20 | 0.09090909 | 0.001375426 | 6.046474 | GCCUAC | GAAUAC,GAGUAC,GCCUGC,GGCUGC,GGGUAC,UAAUAC,UAAUGC,UCAUAC,UCCUGC,UCGUGC,UGAUAC,UGCUGC,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot

Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| IGHMBP2 | 7 | 350 | 0.019047619 | 0.0017684401 | 3.429061 | AAAAAA | AAAAAA |
| SART3 | 13 | 777 | 0.033333333 | 0.0039197904 | 3.088117 | AAAAAA,AAAAAC,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC4 | 7 | 462 | 0.019047619 | 0.0023327287 | 3.029520 | AAAAAA | AAAAAA,AAAAAG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| AGO2 | 10 | 677 | 0.026190476 | 0.0034159613 | 2.938679 | AAAAAA,AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC1 | 15 | 1321 | 0.038095238 | 0.0066606207 | 2.515882 | AAAAAA,AAAAAC,ACAAAU,AGAAAA,CAAACA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| IGF2BP3 | 15 | 1348 | 0.038095238 | 0.0067966546 | 2.486714 | AAAAAC,AAAACA,AAACAC,AAAUCA,AACACA,AAUACA,AAUUCA,ACACAC,CAAACA,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RBM3 | 5 | 541 | 0.014285714 | 0.0027307537 | 2.387202 | AAAACU,AAACUA,AAGACU,AUACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPLL | 7 | 748 | 0.019047619 | 0.0037736800 | 2.335567 | ACACAC,CAAACA,CACACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPL | 14 | 1419 | 0.035714286 | 0.0071543732 | 2.319604 | AAACAA,AAACAC,AAAUAA,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,CACACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| A1CF | 5 | 598 | 0.014285714 | 0.0030179363 | 2.242939 | AGUAUA,AUAAUU,UAAUUG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SRSF4 | 4 | 558 | 0.011904762 | 0.0028164047 | 2.079612 | AAGAAG,AGAAGA,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| IGF2BP2 | 18 | 2163 | 0.045238095 | 0.0109028617 | 2.052831 | AAAAAC,AAAACA,AAACAC,AAAUCA,AACACA,AAUACA,AAUUCA,ACACAC,CAAACA,CAAUUC,CACACA,CCAAAC,GCAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SYNCRIP | 7 | 1041 | 0.019047619 | 0.0052498992 | 1.859249 | AAAAAA | AAAAAA,UUUUUU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | GACUUA,GAUUAA,GCUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| TRA2A | 7 | 1133 | 0.019047619 | 0.0057134220 | 1.737184 | AAAGAA,AAGAAA,AAGAAG,AGAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| HNRNPU | 7 | 1136 | 0.019047619 | 0.0057285369 | 1.733372 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPD | 8 | 1488 | 0.021428571 | 0.0075020153 | 1.514186 | AAAAAA,AAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| DAZAP1 | 16 | 2878 | 0.040476190 | 0.0145052398 | 1.480499 | AAAAAA,AAUUUA,AGGUAA,AGGUUG,AGUAAG,AGUAUA,AGUUUA,AGUUUG,UAGGUU,UAGUAU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACACAC,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM5 | 11 | 2359 | 0.028571429 | 0.0118903668 | 1.264780 | AAAAAA,AAGGUA,AGGGAA,AGGUAA,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAGAAG,AGAAGA,AUAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GCCUAC,UAAUAC,UCCUAC,UGAUAC,UGAUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| FUS | 7 | 1711 | 0.019047619 | 0.0086255542 | 1.142922 | AAAAAA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | AGUAAG,AUACUA,CACUGA,CGCUGA,UGCUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUAAAA,AUUUAC,CUAAAA,CUAAAU,UAAAAA,UAAAAG,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AACAGC,AAGAAG,AGAAGA,GAAGAA,UACAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| PPIE | 39 | 9262 | 0.095238095 | 0.0466696896 | 1.029053 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUUU,AAUAAA,AAUAUU,AAUUUA,AAUUUU,AUAAAA,AUAAUU,AUAUUU,AUUAAA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUU,UAAUUU,UAUAAU,UAUUAA,UUAAAA,UUAAAU,UUAAUU,UUUAAU,UUUUAA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.