circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000138768:+:4:75774675:75812375
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000138768:+:4:75774675:75812375 | ENSG00000138768 | ENST00000264904 | + | 4 | 75774676 | 75812375 | 2265 | GGCGUCUUACUACUGCAGGCACUAACAAGAAGCAAUGGUGCAAUCCAGAAAAUUGUUGCUUUUGAAAAUGCUUUCGAGAGACUACUGGACAUUAUUUCAGAGGAGGGGAACAGUGAUGGAGGUAUAGUAGUUGAAGAUUGUUUGAUUUUGCUCCAAAACUUAUUAAAAAACAACAACUCCAAUCAAAAUUUUUUUAAAGAAGGCUCAUAUAUUCAACGUAUGAAACCUUGGUUUGAAGUUGGAGAUGAAAAUUCUGGCUGGUCUGCACAGAAAGUGACCAAUCUACAUCUAAUGCUACAGCUUGUUCGUGUAUUGGUAUCUCCCACCAACCCUCCUGGUGCUACCAGUAGCUGCCAGAAGGCUAUGUUCCAGUGUGGGUUAUUGCAGCAGCUUUGUACUAUCCUAAUGGCUACUGGGGUUCCUGCUGAUAUCCUGACUGAGACCAUAAAUACUGUAUCAGAAGUUAUUCGAGGCUGCCAAGUAAACCAAGACUACUUUGCAUCUGUAAAUGCACCUUCAAACCCACCAAGACCGGCAAUUGUAGUACUUCUCAUGUCCAUGGUUAAUGAAAGGCAGCCAUUUGUUUUGCGCUGUGCUGUUCUCUAUUGUUUCCAGUGUUUCUUGUAUAAAAACCAAAAAGGACAAGGAGAAAUCGUGUCAACACUUUUACCUUCUACCAUUGAUGCAACAGGUAAUUCAGUUUCAGCUGGCCAGUUAUUAUGUGGAGGUUUGUUUUCUACUGAUUCACUUUCAAACUGGUGUGCUGCUGUGGCCCUUGCCCAUGCGUUGCAAGAAAAUGCCACCCAGAAAGAACAGUUGCUCAGGGUUCAACUUGCUACAAGUAUUGGCAACCCUCCAGUUUCUUUACUUCAACAGUGCACCAAUAUUCUUUCACAGGGUGAUAAGAUCGACAGACGGGGAAGCAAAAUACAAACAAGAGUUGGAUUAUUAAUGUUGCUUUGUACCUGGCUAAGCAAUUGUCCCAUUGCAGUAACGCAUUUUCUUCACAAUUCAGCCAAUGUUCCAUUUCUUACAGGACAAAUUGCAGAAAAUCUUGGAGAAGAAGAGCAGUUGGUCCAAGGCUUAUGUGCCCUUUUGUUGGGCAUUUCGAUUUAUUUCAAUGAUAACUCACUUGAGAGCUACAUGAAAGAGAAGCUAAAACAACUGAUUGAGAAGAGGAUUGGCAAAGAGAAUUUCAUAGAGAAACUAGGAUUUAUUAGCAAACAUGAGUUGUAUUCCAGAGCAUCUCAGAAACCCCAGCCAAACUUUCCCAGUCCAGAAUACAUGAUAUUUGAUCAUGAGUUUACGAAGCUGGUAAAAGAACUUGAAGGUGUUAUAACUAAGGCUAUUUAUAAGUCCAGUGAAGAAGAUAAAAAAGAAGAAGAGGUGAAAAAAACAUUAGAACAGCAUGACAAUAUUGUGACUCACUACAAAAAUAUGAUUCGAGAGCAGGAUCUCCAACUUGAGGAAUUAAGGCAGCAGGUUUCUACAUUAAAAUGUCAAAAUGAACAGCUCCAGACGGCAGUCACACAGCAAGUAUCACAGAUCCAGCAGCACAAAGACCAGUAUAAUCUUCUUAAAAUACAGCUAGGAAAAGACAAUCAGCAUCAAGGUUCUUACAGUGAGGGGGCUCAGAUGAAUGGCAUUCAGCCAGAAGAAAUUGGUAGAUUGCGAGAAGAGAUAGAAGAAUUAAAACGUAAUCAGGAACUUUUACAAAGCCAGCUGACUGAAAAGGACUCUAUGAUUGAAAAUAUGAAAUCUUCCCAAACAUCUGGCACAAAUGAACAGUCUUCAGCAAUAGUUUCAGCUAGAGAUUCUGAACAAGUUGCAGAAUUAAAACAGGAACUGGCAACUUUAAAGUCUCAGUUAAACUCACAAUCUGUGGAGAUCACCAAACUACAGACAGAAAAGCAGGAACUGUUACAGAAAACAGAAGCGUUUGCAAAAUCAGUUGAGGUACAAGGAGAGACCGAGACUAUAAUAGCCACCAAAACUACUGAUGUAGAAGGAAGACUGUCAGCAUUAUUACAAGAGACCAAAGAGUUAAAGAAUGAAAUUAAAGCUCUGUCUGAGGAAAGAACUGCCAUUAAAGAGCAGCUGGAUUCAUCUAAUAGUACCAUUGCCAUUUUACAAACUGAGAAAGACAAACUAGAGUUGGAAAUUACAGAUUCUAAAAAAGAACAAGAUGAUCUCUUGGUGCUCUUGGCCGAUCAAGAUCAGAAAAUACUGUCAUUGAAGAAUAAACUCAAGGAUCUUGGUCAUCCAGGCGUCUUACUACUGCAGGCACUAACAAGAAGCAAUGGUGCAAUCCAGAA | circ |
| ENSG00000138768:+:4:75774675:75812375 | ENSG00000138768 | ENST00000264904 | + | 4 | 75774676 | 75812375 | 22 | UUGGUCAUCCAGGCGUCUUACU | bsj |
| ENSG00000138768:+:4:75774675:75812375 | ENSG00000138768 | ENST00000264904 | + | 4 | 75774476 | 75774685 | 210 | CUAGUGUCUGUCUCUUUGAGGUUUAUAACCUUAGGGAGACAAAUUGCUUAACUAUUCUGUUCUUGUUUUCCUUAUGUAUAAAGUAAAGGAGUUGGGCUUGCAAAAUCUCUGCCAGUUCUAAAAUUCUGUGAUUUUUGAAGUAAUUUAAAAAUGUCUCAUAAAUUUUGUACUCCACUAAACAUUCUCUUUCUUCUCUAUAGGGCGUCUUAC | ie_up |
| ENSG00000138768:+:4:75774675:75812375 | ENSG00000138768 | ENST00000264904 | + | 4 | 75812366 | 75812575 | 210 | UGGUCAUCCAGUAAGAUUAAAAUGUCUCCAAAAAUGAUUUGUAUUAUGUUUUAGUAUAAUGCAUUUAAAAGAUUUUUAAUGUAACAUGGAAAAGUUGUGCCUGUAUUAUAGGAAUGGAUAUGUGGGUUCAUGAAUAUGGUACCAUCUAUACUGUUUUCACUGUUUUCAUUGAAUUUACUUAAUACACAGAUUAGGUUCAGUUUUCAGUAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
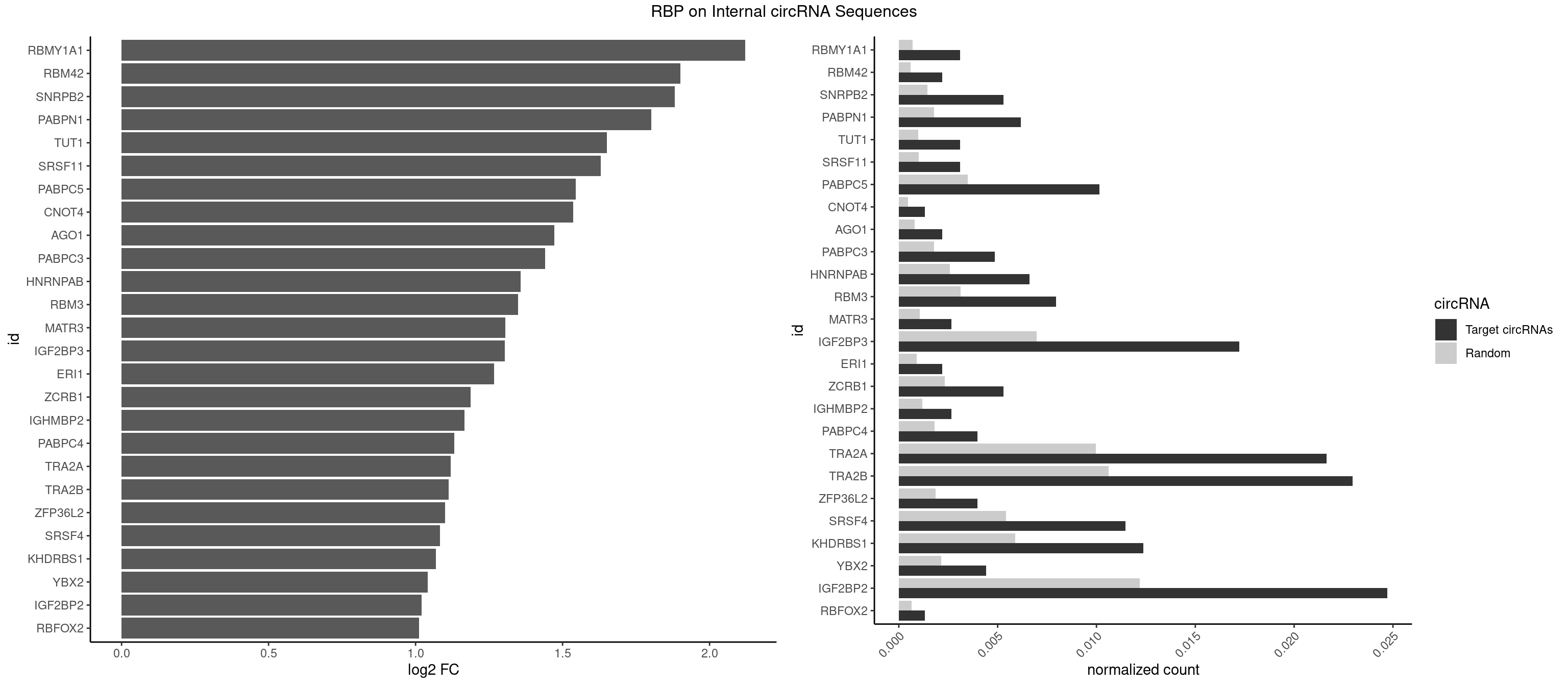
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 6 | 489 | 0.003090508 | 0.0007101099 | 2.121730 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| RBM42 | 4 | 407 | 0.002207506 | 0.0005912752 | 1.900515 | AACUAA,AACUAC,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| SNRPB2 | 11 | 991 | 0.005298013 | 0.0014376103 | 1.881779 | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPN1 | 13 | 1222 | 0.006181015 | 0.0017723764 | 1.802159 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| TUT1 | 6 | 678 | 0.003090508 | 0.0009840095 | 1.651100 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF11 | 6 | 688 | 0.003090508 | 0.0009985015 | 1.630007 | AAGAAG | AAGAAG |
| PABPC5 | 22 | 2400 | 0.010154525 | 0.0034795387 | 1.545155 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CNOT4 | 2 | 314 | 0.001324503 | 0.0004564992 | 1.536767 | GACAGA | GACAGA |
| AGO1 | 4 | 548 | 0.002207506 | 0.0007956130 | 1.472278 | GAGGUA,GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPC3 | 10 | 1234 | 0.004856512 | 0.0017897669 | 1.440149 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPAB | 14 | 1782 | 0.006622517 | 0.0025839306 | 1.357812 | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 17 | 2152 | 0.007947020 | 0.0031201361 | 1.348805 | AAAACG,AAAACU,AAACUA,AAGACU,AAUACU,AGACUA,GAAACU,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| MATR3 | 5 | 739 | 0.002649007 | 0.0010724109 | 1.304594 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IGF2BP3 | 38 | 4815 | 0.017218543 | 0.0069793662 | 1.302795 | AAAAAC,AAAACA,AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,CAAACA,CAAUCA,CACACA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ERI1 | 4 | 632 | 0.002207506 | 0.0009173461 | 1.266879 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| ZCRB1 | 11 | 1605 | 0.005298013 | 0.0023274215 | 1.186719 | AAUUAA,GAAUUA,GAUUUA,GGAUUA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGHMBP2 | 5 | 813 | 0.002649007 | 0.0011796520 | 1.167090 | AAAAAA | AAAAAA |
| PABPC4 | 8 | 1251 | 0.003973510 | 0.0018144033 | 1.130919 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| TRA2A | 48 | 6871 | 0.021633554 | 0.0099589296 | 1.119208 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TRA2B | 51 | 7329 | 0.022958057 | 0.0106226650 | 1.111855 | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ZFP36L2 | 8 | 1277 | 0.003973510 | 0.0018520827 | 1.101265 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| SRSF4 | 25 | 3740 | 0.011479029 | 0.0054214720 | 1.082244 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| KHDRBS1 | 27 | 4064 | 0.012362031 | 0.0058910141 | 1.069328 | AUAAAA,CAAAAU,CUAAAA,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YBX2 | 9 | 1480 | 0.004415011 | 0.0021462711 | 1.040585 | AACAAC,AACAUC,ACAACA,ACAACU,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| IGF2BP2 | 55 | 8408 | 0.024724062 | 0.0121863560 | 1.020649 | AAAAAC,AAAACA,AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,CAAAAC,CAAACA,CAACAC,CAACUC,CAAUCA,CAAUUC,CACACA,CAUUCA,CCAAAC,CCAAUC,GAAAAC,GAAAUC,GAAUAC,GCAAAC,GCAAUC,GCAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBFOX2 | 2 | 452 | 0.001324503 | 0.0006564894 | 1.012608 | UGACUG | UGACUG,UGCAUG |
RBP on BSJ (Exon Only)
Plot
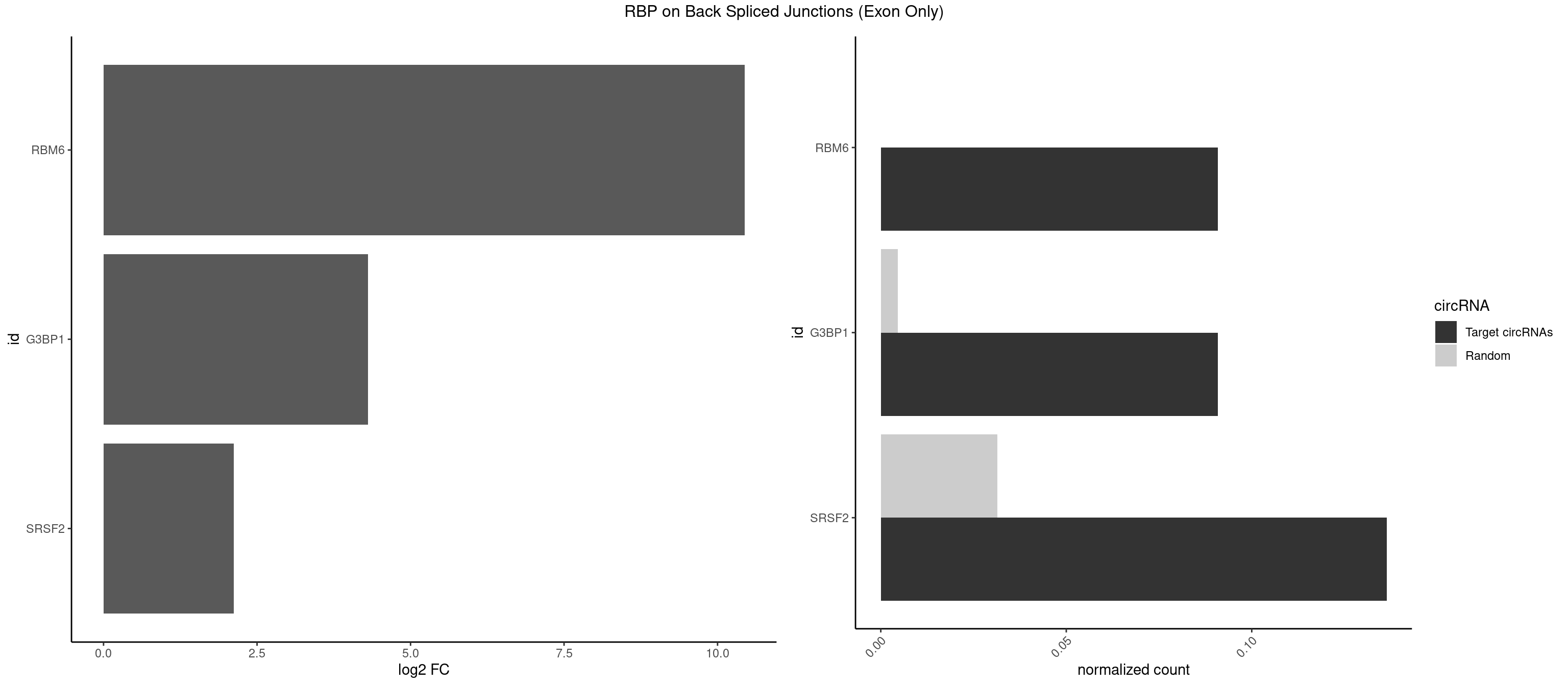
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM6 | 1 | 0 | 0.09090909 | 6.549646e-05 | 10.438792 | AUCCAG | |
| G3BP1 | 1 | 69 | 0.09090909 | 4.584752e-03 | 4.309509 | CCAGGC | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| SRSF2 | 2 | 479 | 0.13636364 | 3.143830e-02 | 2.116864 | AGGCGU,UCCAGG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
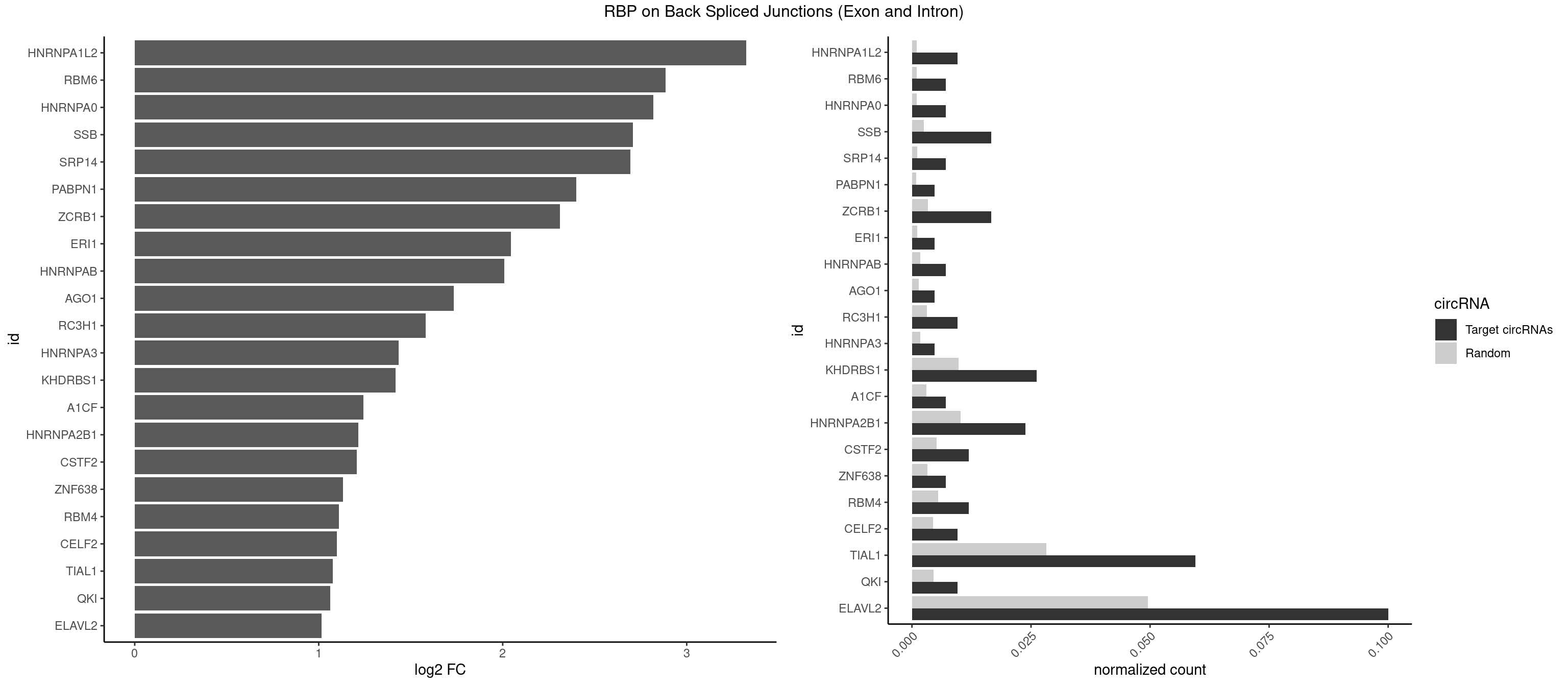
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 3 | 188 | 0.009523810 | 0.0009522370 | 3.322146 | AUAGGG,UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SSB | 6 | 505 | 0.016666667 | 0.0025493753 | 2.708750 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| ZCRB1 | 6 | 666 | 0.016666667 | 0.0033605401 | 2.310201 | ACUUAA,AUUUAA,GAUUAA,GCUUAA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| KHDRBS1 | 10 | 1945 | 0.026190476 | 0.0098045143 | 1.417524 | AUUUAA,AUUUAC,CAAAAU,CUAAAA,UAAAAA,UAAAAG,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AGUAUA,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPA2B1 | 9 | 2033 | 0.023809524 | 0.0102478839 | 1.216213 | AAGGAG,AAUUUA,AUAGGG,AUUUAA,UAGGGA,UUAGGG,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CUCUUU,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUCUGU,UAUGUG,UAUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AAAUUU,AAUUUU,AGUUUU,AUUUUG,AUUUUU,CAGUUU,GUUUUC,UAAAUU,UCAGUU,UUCAGU,UUUAAA,UUUCAG,UUUUAA,UUUUCA,UUUUUA,UUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | CACUAA,CUCAUA,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ELAVL2 | 41 | 9826 | 0.100000000 | 0.0495112858 | 1.014171 | AAUUUA,AAUUUU,AUUUAA,AUUUAC,AUUUUG,AUUUUU,CUUUCU,GAUUUU,GUUUUA,GUUUUC,UAAUUU,UAUACU,UAUGUU,UAUUAU,UGAUUU,UGUAUU,UUACUU,UUAUGU,UUCAUU,UUGUAU,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUCAU,UUUCUU,UUUUAA,UUUUAG,UUUUUA,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.