circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000215158:-:5:34179012:34182867
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000215158:-:5:34179012:34182867 | ENSG00000215158 | ENST00000514048 | - | 5 | 34179013 | 34182867 | 273 | GGCUGGUGAAUUACCAUAUCUCCGUCAAGUGCAGUAACCAGUUCAAGUUGGAAGUGUGUCUUUUGAAAUCAGACAACAAAGUCGUGGACAACCAGGCUGGGACCCAGGGCCAGCUGAAGGUGCUGGGUGCCAACCUCUGGUGGCCGUACCUGAUGCACGAACACCCCGCCUACCUGUACUCGUGGGAGGAUGGUGAUUGCUCACACCAAAGCCUUGGACCCCUCCCAGCCUGUGACCUUUGUGACCAACUCCACCUACGCAGCAGACAAGGGGGGCUGGUGAAUUACCAUAUCUCCGUCAAGUGCAGUAACCAGUUCAAGUUG | circ |
| ENSG00000215158:-:5:34179012:34182867 | ENSG00000215158 | ENST00000514048 | - | 5 | 34179013 | 34182867 | 22 | CAGACAAGGGGGGCUGGUGAAU | bsj |
| ENSG00000215158:-:5:34179012:34182867 | ENSG00000215158 | ENST00000514048 | - | 5 | 34182858 | 34183067 | 210 | GGCAGAGGUUUCAGUGAGUCCAGAUCAUGCCACUGCACUCCAGCCUGGGUGACAGAGCGAGAUUCUAUCUCAAAAAAAAAAAAAAAAAAAAGCAACAGAAGCAAAUGAGAGUGCCUGGGAGUGGUCAUUGUGGGGCAUUCCUGUUCGUGUGACCCAGGUCAUGUCCCUCCCUAAGCCCUGGUCUCUCUUGCCUCCUGCAGGGCUGGUGAA | ie_up |
| ENSG00000215158:-:5:34179012:34182867 | ENSG00000215158 | ENST00000514048 | - | 5 | 34178813 | 34179022 | 210 | AGACAAGGGGGUGAGCCUGGGGGUCCCCACCCCAUUUCUCCCUGCCUUUGCCUGGGCUUGUCCUGAAGCCUGCUCAUGGGAACAGCUGGAAAGAACCAUGUGCUGCCAGUCUGAGCUUUUUAUUUUGUUUUACUUAGAAAGAUAGAGACAGGGUCUUGCCAUGUUGCCCAGGCUGGUCUCGAACUCCUGGGCUCAAGUGAUCCUCCUGCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
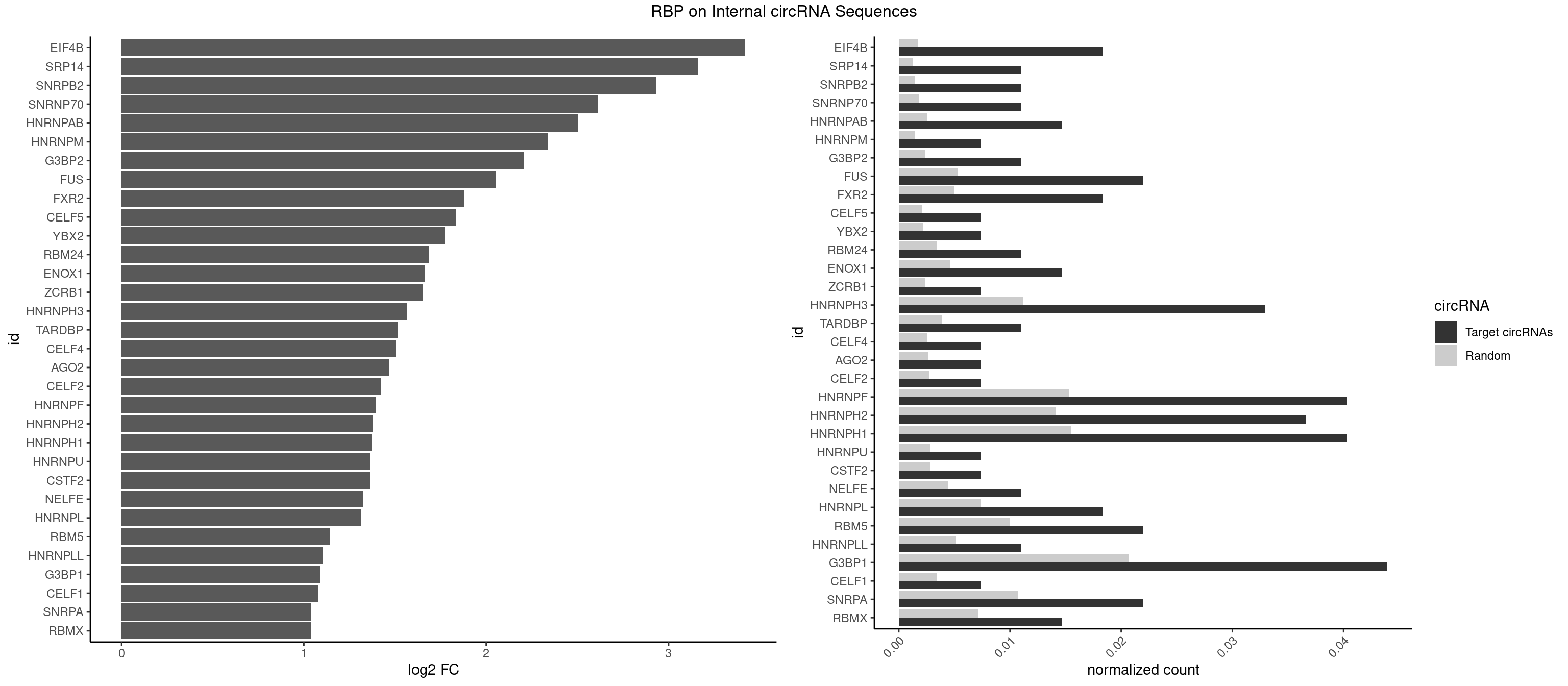
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| EIF4B | 4 | 1179 | 0.018315018 | 0.001710061 | 3.420908 | CUUGGA,GUUGGA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SRP14 | 2 | 847 | 0.010989011 | 0.001228925 | 3.160593 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SNRPB2 | 2 | 991 | 0.010989011 | 0.001437610 | 2.934317 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| SNRNP70 | 2 | 1237 | 0.010989011 | 0.001794114 | 2.614718 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPAB | 3 | 1782 | 0.014652015 | 0.002583931 | 2.503460 | ACAAAG,AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPM | 1 | 999 | 0.007326007 | 0.001449204 | 2.337766 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 2 | 1644 | 0.010989011 | 0.002383941 | 2.204641 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| FUS | 5 | 3648 | 0.021978022 | 0.005288145 | 2.055228 | GGGGGG,GGGUGC,UGGUGA,UGGUGG | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| FXR2 | 4 | 3434 | 0.018315018 | 0.004978016 | 1.879384 | AGACAA,GACAAG,GGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CELF5 | 1 | 1415 | 0.007326007 | 0.002052073 | 1.835945 | GUGUGU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 1 | 1480 | 0.007326007 | 0.002146271 | 1.771195 | ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM24 | 2 | 2357 | 0.010989011 | 0.003417223 | 1.685165 | AGUGUG,GUGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ENOX1 | 3 | 3195 | 0.014652015 | 0.004631656 | 1.661499 | CAGACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZCRB1 | 1 | 1605 | 0.007326007 | 0.002327422 | 1.654295 | GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPH3 | 8 | 7688 | 0.032967033 | 0.011142929 | 1.564896 | AAGGUG,GGAGGA,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,UGGGUG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| TARDBP | 2 | 2654 | 0.010989011 | 0.003847636 | 1.514017 | GUGAAU,GUGUGU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF4 | 1 | 1782 | 0.007326007 | 0.002583931 | 1.503460 | GUGUGU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO2 | 1 | 1830 | 0.007326007 | 0.002653492 | 1.465135 | AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CELF2 | 1 | 1886 | 0.007326007 | 0.002734648 | 1.421672 | GUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| HNRNPF | 10 | 10561 | 0.040293040 | 0.015306492 | 1.396387 | AAGGGG,AAGGUG,GAAGGU,GGAGGA,GGAUGG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH2 | 9 | 9711 | 0.036630037 | 0.014074669 | 1.379926 | AAGGGG,AAGGUG,GGAGGA,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 10 | 10717 | 0.040293040 | 0.015532568 | 1.375234 | AAGGGG,AAGGUG,GAAGGU,GGAGGA,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPU | 1 | 1965 | 0.007326007 | 0.002849135 | 1.362503 | GGGGGG | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| CSTF2 | 1 | 1967 | 0.007326007 | 0.002852033 | 1.361036 | GUGUGU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| NELFE | 2 | 3028 | 0.010989011 | 0.004389639 | 1.323887 | CUCUGG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| HNRNPL | 4 | 5085 | 0.018315018 | 0.007370651 | 1.313163 | AACAAA,ACACCA,CACACC,CACGAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBM5 | 5 | 6879 | 0.021978022 | 0.009970523 | 1.140320 | AAGGGG,AAGGUG,CAAGGG,GAAGGU,GGGGGG | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPLL | 2 | 3534 | 0.010989011 | 0.005122936 | 1.101019 | ACACCA,CACACC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| G3BP1 | 11 | 14282 | 0.043956044 | 0.020698980 | 1.086502 | ACCCCU,ACGCAG,CCAGGC,CCCAGG,CCCCGC,CCCCUC,CCCGCC,CCCUCC,CCUACG,CUACGC,UACGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| CELF1 | 1 | 2391 | 0.007326007 | 0.003466496 | 1.079549 | GUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SNRPA | 5 | 7380 | 0.021978022 | 0.010696574 | 1.038913 | AUGCAC,GCAGUA,UACCUG,UGCACG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RBMX | 3 | 4925 | 0.014652015 | 0.007138779 | 1.037350 | AAGUGU,ACCAAA,AGUAAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
RBP on BSJ (Exon Only)
Plot
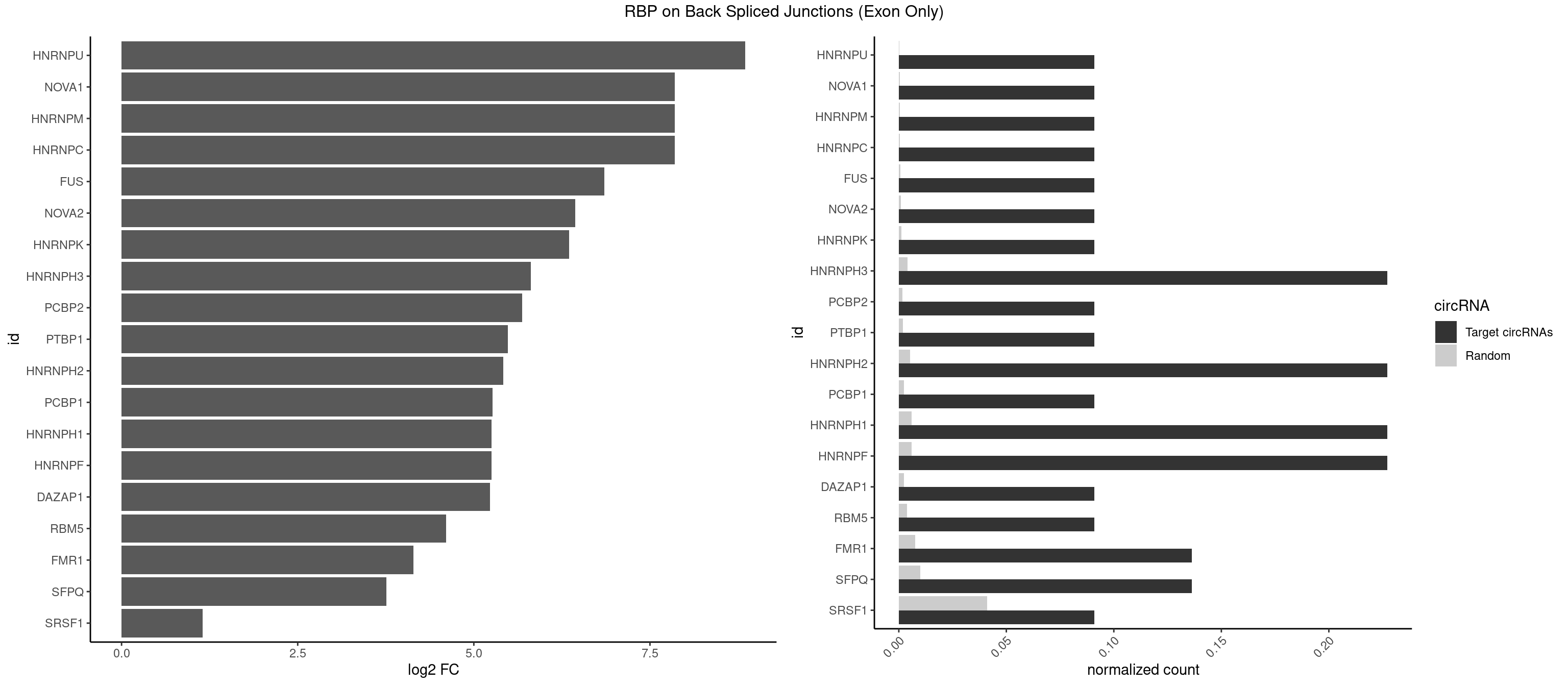
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPU | 1 | 2 | 0.09090909 | 0.0001964894 | 8.853829 | GGGGGG | AAAAAA |
| HNRNPC | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GGGGGG | GGAUAU,GGAUUC,GGGUAC |
| HNRNPM | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GGGGGG | AAGGAA,GAAGGA |
| NOVA1 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GGGGGG | ACCACC,CAGUCA,CAUUCA,UCAGUC |
| FUS | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | GGGGGG | AAAAAA,CGGUGG,GGGUGA,GGGUGG,GGGUGU |
| NOVA2 | 1 | 15 | 0.09090909 | 0.0010479434 | 6.438792 | GGGGGG | ACCACC,AGACAU,AGAUCA,AGGCAU,AGUCAU,CUAGAU,GAGACA,GAGGCA,GAGUCA,UAGAUC |
| HNRNPK | 1 | 16 | 0.09090909 | 0.0011134399 | 6.351329 | GGGGGG | AAAAAA,ACCCCA,ACCCCC,ACCCGA,CAAACC,CAUACC,CCAACC,CCAUAC,CCCCAC,CCCCCU,CCCCUU,GCCCAC,GCCCAG,UCCCAC |
| HNRNPH3 | 4 | 61 | 0.22727273 | 0.0040607807 | 5.806524 | GGGCUG,GGGGCU,GGGGGC,GGGGGG | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| PCBP2 | 1 | 26 | 0.09090909 | 0.0017684045 | 5.683904 | GGGGGG | AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,AUUAAA,CCAUUC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CUCCCU,CUUCCA,UAACCC,UUAGAG,UUAGGA,UUAGGG |
| PTBP1 | 1 | 30 | 0.09090909 | 0.0020303904 | 5.484596 | GGGGGG | AGCUGU,CAUCUU,CCAUCU,CCUCUU,CUAUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUUCU,GGCUCC,GUCUUU,UACUUU,UAUUUU,UCCUCU,UCUAUC,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUCUUC,UUCUUG,UUUCUU |
| HNRNPH2 | 4 | 80 | 0.22727273 | 0.0053052135 | 5.420870 | GGGCUG,GGGGCU,GGGGGC,GGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| PCBP1 | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | GGGGGG | AAAAAA,AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,ACCCUC,AUUAAA,CAAACC,CAUACC,CCAACC,CCACCC,CCAUAC,CCAUUC,CCCACC,CCCCAC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,UUAGAG,UUAGGA,UUAGGG |
| HNRNPF | 4 | 90 | 0.22727273 | 0.0059601782 | 5.252925 | GGGCUG,GGGGCU,GGGGGC,GGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 4 | 90 | 0.22727273 | 0.0059601782 | 5.252925 | GGGCUG,GGGGCU,GGGGGC,GGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | GGGGGG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| RBM5 | 1 | 56 | 0.09090909 | 0.0037332984 | 4.605902 | GGGGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| FMR1 | 2 | 117 | 0.13636364 | 0.0077285827 | 4.141111 | GGGCUG,GGGGGG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SFPQ | 2 | 153 | 0.13636364 | 0.0100864553 | 3.756968 | AGGGGG,GGGGGG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF1 | 1 | 625 | 0.09090909 | 0.0410007860 | 1.148773 | GGGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
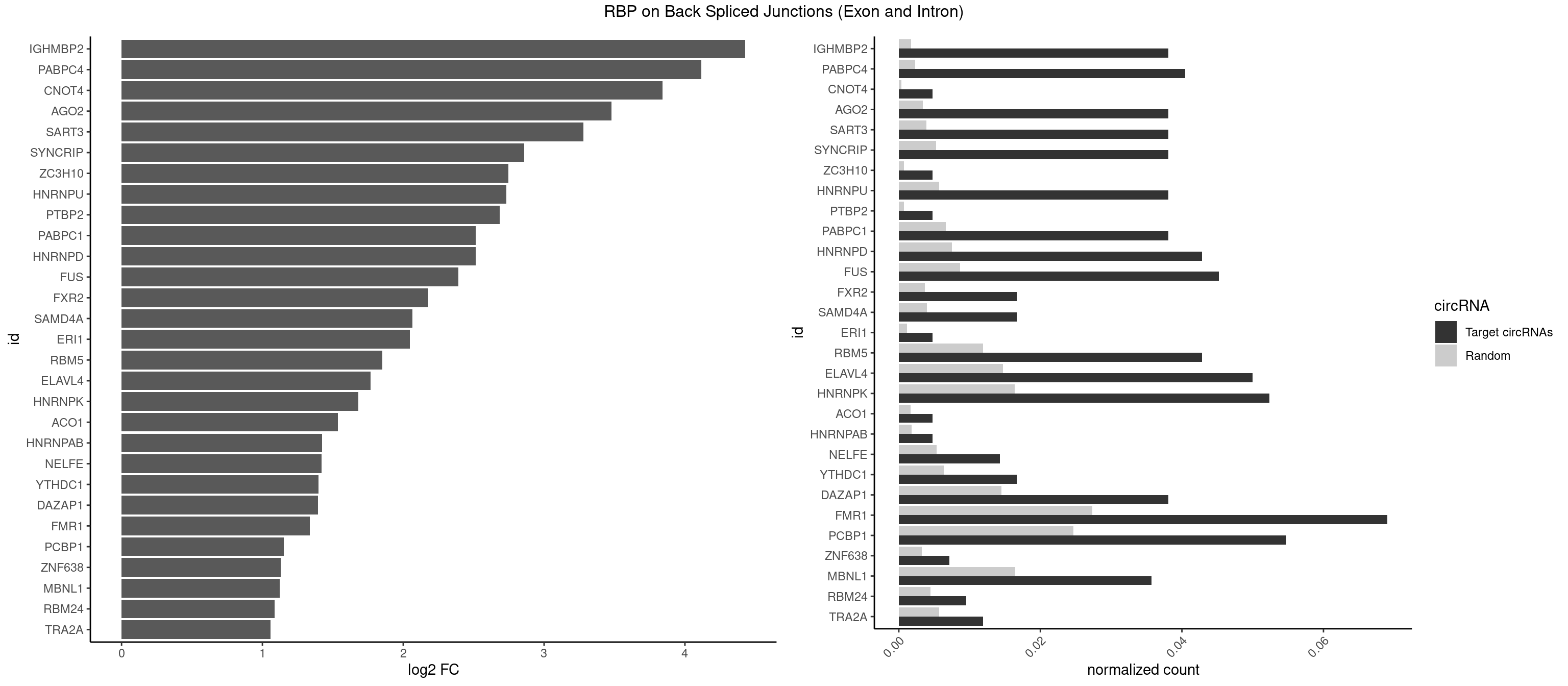
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 15 | 350 | 0.038095238 | 0.0017684401 | 4.429061 | AAAAAA | AAAAAA |
| PABPC4 | 16 | 462 | 0.040476190 | 0.0023327287 | 4.116983 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| AGO2 | 15 | 677 | 0.038095238 | 0.0034159613 | 3.479247 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SART3 | 15 | 777 | 0.038095238 | 0.0039197904 | 3.280762 | AAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SYNCRIP | 15 | 1041 | 0.038095238 | 0.0052498992 | 2.859249 | AAAAAA | AAAAAA,UUUUUU |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| HNRNPU | 15 | 1136 | 0.038095238 | 0.0057285369 | 2.733372 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| PABPC1 | 15 | 1321 | 0.038095238 | 0.0066606207 | 2.515882 | AAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPD | 17 | 1488 | 0.042857143 | 0.0075020153 | 2.514186 | AAAAAA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| FUS | 18 | 1711 | 0.045238095 | 0.0086255542 | 2.390849 | AAAAAA,GGGUGA,UGGUGA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| FXR2 | 6 | 730 | 0.016666667 | 0.0036829907 | 2.178016 | AGACAA,AGACAG,GACAAG,GACAGA,GACAGG,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SAMD4A | 6 | 790 | 0.016666667 | 0.0039852882 | 2.064210 | CUGGAA,CUGGUC,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| RBM5 | 17 | 2359 | 0.042857143 | 0.0118903668 | 1.849742 | AAAAAA,AAGGGG,CAAGGG | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| ELAVL4 | 20 | 2916 | 0.050000000 | 0.0146966949 | 1.766436 | AAAAAA,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPK | 21 | 3244 | 0.052380952 | 0.0163492543 | 1.679817 | AAAAAA,ACCCCA,CCCCAC,CCCCAU,GCCCAG,UCCCCA,UCCCUA | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUCU,GGUCUC,GUCUCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.0063230552 | 1.398272 | GAGUGC,GCCUGC,UCAUGC,UCCUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| DAZAP1 | 15 | 2878 | 0.038095238 | 0.0145052398 | 1.393037 | AAAAAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| FMR1 | 28 | 5430 | 0.069047619 | 0.0273629585 | 1.335367 | AAAAAA,AGCUGG,CAGCUG,CUGGGC,CUGGUG,GACAAG,GACAGG,GAGCGA,GGCUGG,GGGCAU,GGGCUG,UGACAG | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| PCBP1 | 22 | 4891 | 0.054761905 | 0.0246473196 | 1.151742 | AAAAAA,CAUUCC,CCACCC,CCCACC,CCCCAC,CCCUCC,CCUCCC,UCCCCA | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCGU,UGUUCG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| MBNL1 | 14 | 3258 | 0.035714286 | 0.0164197904 | 1.121066 | AUGCCA,CCUGCU,CUGCCA,CUGCCU,CUGCUC,GCUUGU,GCUUUU,GUCUCG,GUGCUG,UGCUGC,UUGCCA,UUGCCC,UUGCCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | AGAGUG,GAGUGG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAAGAA,AGAAAG,GAAAGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.