circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000044115:+:5:138781922:138783372
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000044115:+:5:138781922:138783372 | ENSG00000044115 | ENST00000627109 | + | 5 | 138781923 | 138783372 | 303 | AAAUGACUGCUGUCCAUGCAGGCAACAUAAACUUCAAGUGGGAUCCUAAAAGUCUAGAGAUCAGGACUCUGGCAGUUGAGAGACUGUUGGAGCCUCUUGUUACACAGGUUACAACCCUUGUAAACACCAAUAGUAAAGGGCCCUCUAAUAAGAAGAGAGGUCGUUCUAAGAAGGCCCAUGUUUUGGCUGCAUCUGUUGAACAAGCAACUGAGAAUUUCUUGGAGAAGGGGGAUAAAAUUGCGAAGGAGAGCCAGUUUCUCAAGGAGGAGCUUGUGGCUGCUGUAGAAGAUGUUCGAAAACAAGAAAUGACUGCUGUCCAUGCAGGCAACAUAAACUUCAAGUGGGAUCCUAAA | circ |
| ENSG00000044115:+:5:138781922:138783372 | ENSG00000044115 | ENST00000627109 | + | 5 | 138781923 | 138783372 | 22 | UCGAAAACAAGAAAUGACUGCU | bsj |
| ENSG00000044115:+:5:138781922:138783372 | ENSG00000044115 | ENST00000627109 | + | 5 | 138781723 | 138781932 | 210 | CCUAUUUGUAAGCCUGAAAUCAUUAAAUAUUAAGAGGUUGUAAGGUUUACUGGGUCUUCAUGUGUAGGCUAGUUAGAAUUUUUUCAUCUAUAGUGAAUAUAGCUUUCCUGAUGCAAAAGUCCCAAAGUAGGAGAAGAUUUGUUACAUAUUUCAUGUUUGUGUUUGAUGUUUGCCUGACUGACUUUUUGUUUCUUAUUUAGAAAUGACUGC | ie_up |
| ENSG00000044115:+:5:138781922:138783372 | ENSG00000044115 | ENST00000627109 | + | 5 | 138783363 | 138783572 | 210 | CGAAAACAAGGUAGGUCAUUACUGCUUUUUAGGUAAAGAGAGGCAGGCCUUUCUAGAAAAUCAGUGUUUGAAGAUUUUUUUUGUGGUCAGUAUUCUCAUUCUUAUGCUUGCCAAUUGCUCAGUAACUUACUUAGUUAUUGGAGAUGUAUUAAGUUGGACUGGCUCAUAUUUUAAUUCUCAUUCUUAUGCUUGCCAGUUGCUCAGUAACUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
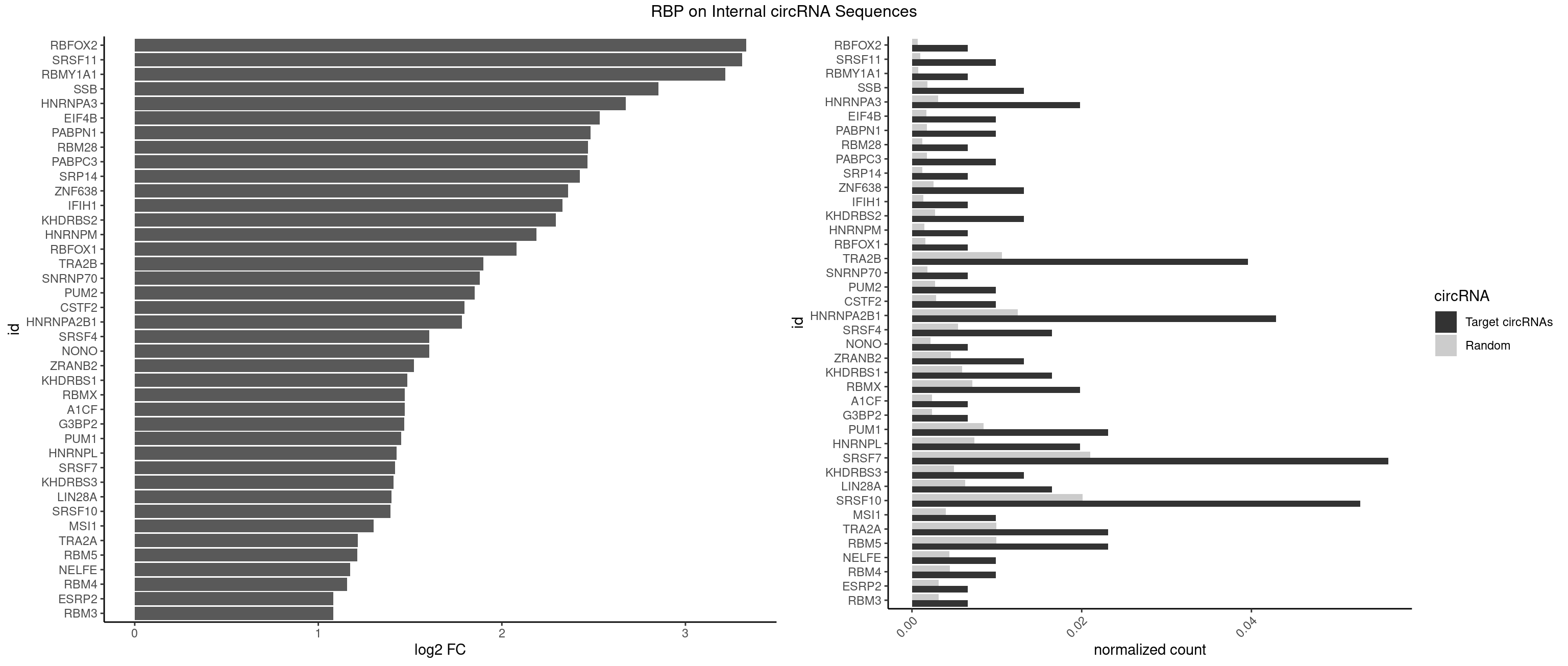
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 452 | 0.00660066 | 0.0006564894 | 3.329767 | UGACUG | UGACUG,UGCAUG |
| SRSF11 | 2 | 688 | 0.00990099 | 0.0009985015 | 3.309736 | AAGAAG | AAGAAG |
| RBMY1A1 | 1 | 489 | 0.00660066 | 0.0007101099 | 3.216496 | ACAAGA | ACAAGA,CAAGAC |
| SSB | 3 | 1260 | 0.01320132 | 0.0018274462 | 2.852781 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPA3 | 5 | 2140 | 0.01980198 | 0.0031027457 | 2.674027 | AAGGAG,AGGAGC,CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| EIF4B | 2 | 1179 | 0.00990099 | 0.0017100607 | 2.533525 | CUUGGA,GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPN1 | 2 | 1222 | 0.00990099 | 0.0017723764 | 2.481888 | AGAAGA | AAAAGA,AGAAGA |
| RBM28 | 1 | 822 | 0.00660066 | 0.0011926949 | 2.468385 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPC3 | 2 | 1234 | 0.00990099 | 0.0017897669 | 2.467801 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRP14 | 1 | 847 | 0.00660066 | 0.0012289250 | 2.425213 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ZNF638 | 3 | 1773 | 0.01320132 | 0.0025708878 | 2.360344 | CGUUCU,UGUUCG,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| IFIH1 | 1 | 904 | 0.00660066 | 0.0013115296 | 2.331360 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| KHDRBS2 | 3 | 1858 | 0.01320132 | 0.0026940701 | 2.292823 | AUAAAA,AUAAAC,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPM | 1 | 999 | 0.00660066 | 0.0014492040 | 2.187350 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBFOX1 | 1 | 1077 | 0.00660066 | 0.0015622419 | 2.078992 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TRA2B | 11 | 7329 | 0.03960396 | 0.0106226650 | 1.898499 | AAGAAG,AAUAAG,AGAAGA,AGAAGG,AUAAGA,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SNRNP70 | 1 | 1237 | 0.00660066 | 0.0017941145 | 1.879338 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 2 | 1890 | 0.00990099 | 0.0027404447 | 1.853163 | UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CSTF2 | 2 | 1967 | 0.00990099 | 0.0028520334 | 1.795582 | GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPA2B1 | 12 | 8607 | 0.04290429 | 0.0124747476 | 1.782111 | AAGAAG,AAGGAG,AAGGGG,AGGAGC,CAAGAA,CAAGGA,CGAAGG,GAAGGA,GCGAAG,GGAGCC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| SRSF4 | 4 | 3740 | 0.01650165 | 0.0054214720 | 1.605854 | AAGAAG,AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO | 1 | 1498 | 0.00660066 | 0.0021723567 | 1.603349 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ZRANB2 | 3 | 3173 | 0.01320132 | 0.0045997733 | 1.521048 | AGGUUA,GUAAAG,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS1 | 4 | 4064 | 0.01650165 | 0.0058910141 | 1.486022 | AUAAAA,CUAAAA,GAAAAC,UAAAAG | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBMX | 5 | 4925 | 0.01980198 | 0.0071387787 | 1.471896 | AAGAAG,AGAAGG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| A1CF | 1 | 1642 | 0.00660066 | 0.0023810421 | 1.471017 | GAUCAG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP2 | 1 | 1644 | 0.00660066 | 0.0023839405 | 1.469262 | GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PUM1 | 6 | 5823 | 0.02310231 | 0.0084401638 | 1.452694 | ACAUAA,AGAAUU,CUUGUA,UGUAAA,UGUAGA,UGUCCA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPL | 5 | 5085 | 0.01980198 | 0.0073706513 | 1.425781 | AAACAA,AAACAC,ACACCA,ACAUAA,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| SRSF7 | 16 | 14481 | 0.05610561 | 0.0209873716 | 1.418624 | AAGAAG,AGAAGA,AGAGAC,AGAGAG,AGAGAU,AGAUCA,CUAGAG,CUGAGA,GAAGGC,GAGACU,GAGAGA,GAGAUC,UAGAGA,UGAGAG | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| KHDRBS3 | 3 | 3429 | 0.01320132 | 0.0049707696 | 1.409141 | AUAAAA,AUAAAC,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| LIN28A | 4 | 4315 | 0.01650165 | 0.0062547643 | 1.399583 | AGGAGA,GGAGAA,GGAGGA,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF10 | 15 | 13860 | 0.05280528 | 0.0200874160 | 1.394390 | AAAGGG,AAGAAA,AAGAAG,AAGAGA,AAGGAG,AAGGGG,AGAGAC,AGAGAG,GAGAAG,GAGAGA,GAGAGC,GAGAGG,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| MSI1 | 2 | 2770 | 0.00990099 | 0.0040157442 | 1.301905 | AGGAGG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| TRA2A | 6 | 6871 | 0.02310231 | 0.0099589296 | 1.213975 | AAGAAA,AAGAAG,AGAAGA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM5 | 6 | 6879 | 0.02310231 | 0.0099705232 | 1.212296 | AAGGAG,AAGGGG,CAAGGA,GAAGGA,GAAGGG | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| NELFE | 2 | 3028 | 0.00990099 | 0.0043896388 | 1.173471 | CUCUGG,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 2 | 3065 | 0.00990099 | 0.0044432593 | 1.155954 | CCUCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ESRP2 | 1 | 2150 | 0.00660066 | 0.0031172377 | 1.082342 | GGGGAU | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM3 | 1 | 2152 | 0.00660066 | 0.0031201361 | 1.081001 | GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
RBP on BSJ (Exon Only)
Plot
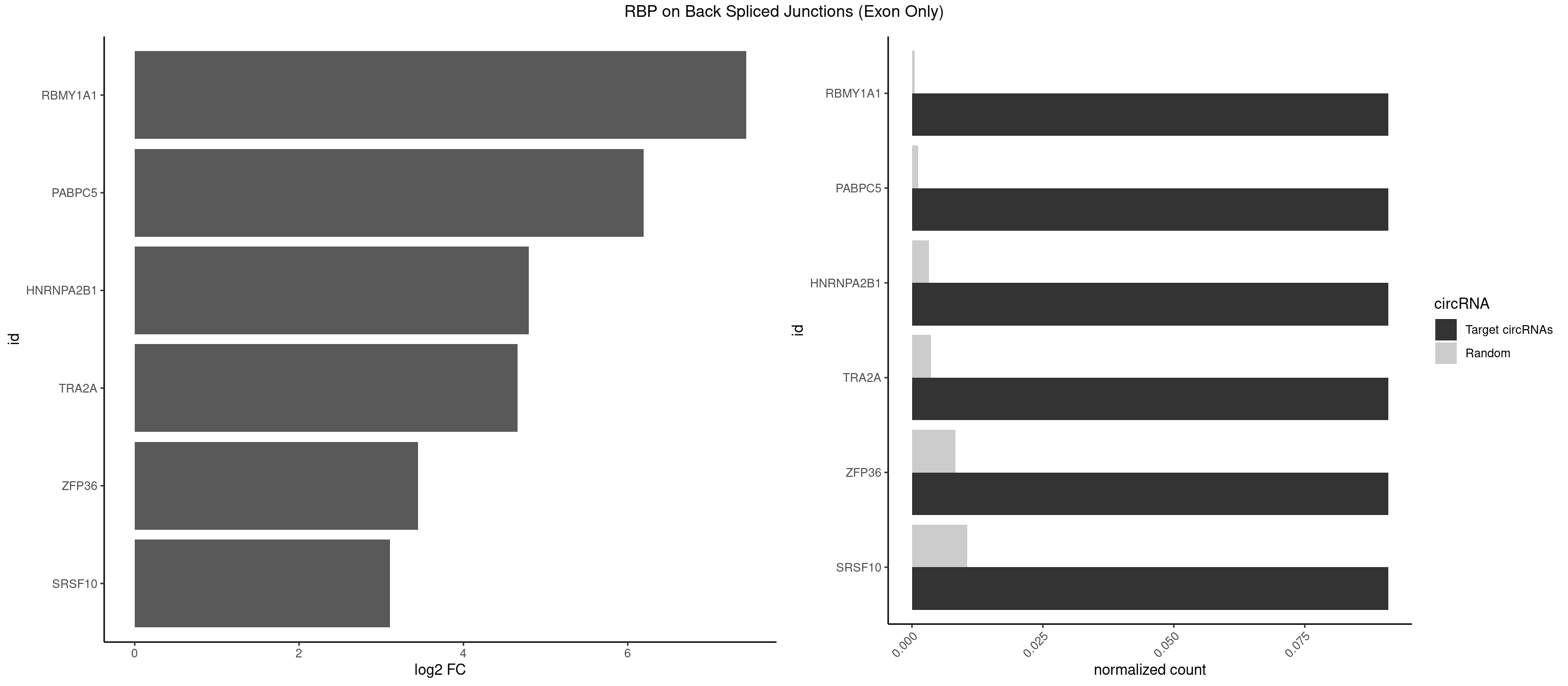
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | ACAAGA | ACAAGA,CAAGAC |
| PABPC5 | 1 | 18 | 0.09090909 | 0.0012444328 | 6.190864 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | CAAGAA | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| TRA2A | 1 | 54 | 0.09090909 | 0.0036023055 | 4.657432 | AAGAAA | AAAGAA,AAGAAA,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | AAGAAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | AAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
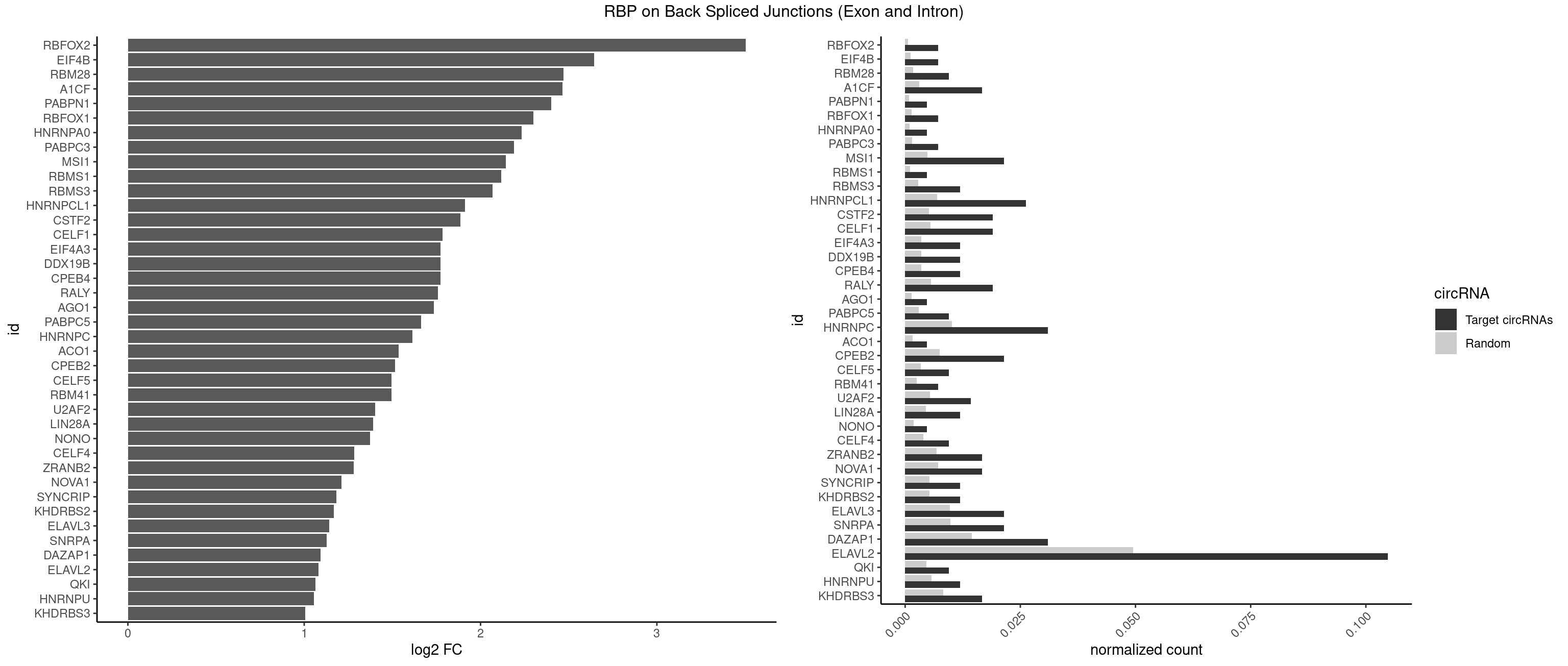
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGACUG | UGACUG,UGCAUG |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 3 | 340 | 0.009523810 | 0.0017180572 | 2.470761 | AGUAGG,GUGUAG,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| A1CF | 6 | 598 | 0.016666667 | 0.0030179363 | 2.465331 | AUCAGU,CAGUAU,UCAGUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| MSI1 | 8 | 961 | 0.021428571 | 0.0048468360 | 2.144421 | AGGUAG,AGUAGG,AGUUAG,AGUUGG,UAGGAG,UAGGUA,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 | 4 | 562 | 0.011904762 | 0.0028365578 | 2.069326 | AAUAUA,AUAUAG,CUAUAG,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPCL1 | 10 | 1381 | 0.026190476 | 0.0069629182 | 1.911278 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF1 | 7 | 1097 | 0.019047619 | 0.0055320435 | 1.783726 | GUUUGU,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CPEB4 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| DDX19B | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| RALY | 7 | 1117 | 0.019047619 | 0.0056328094 | 1.757684 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,AGAAAU,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPC | 12 | 2006 | 0.030952381 | 0.0101118501 | 1.614003 | AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.0054010480 | 1.403262 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGA,GGAGAA,GGAGAU,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ZRANB2 | 6 | 1360 | 0.016666667 | 0.0068571141 | 1.281292 | AGGUAA,AGGUAG,AGGUUU,GAGGUU,GGUAAA,GUAAAG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | UCAUUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.0052498992 | 1.181177 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | AGGAGA,AGUAGG,GGAGAU,GUAGGC,UAUGCU,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| DAZAP1 | 12 | 2878 | 0.030952381 | 0.0145052398 | 1.093476 | AGGUAA,AGGUAG,AGGUUG,AGUAGG,AGUUAG,UAGGUA,UAGUUA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL2 | 43 | 9826 | 0.104761905 | 0.0495112858 | 1.081285 | AAUUUU,AUAUUU,AUUUAG,AUUUUA,AUUUUU,CUUUCU,CUUUUU,GAUUUU,UAAGUU,UAGUUA,UAUUUA,UAUUUG,UAUUUU,UCUUAU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUUG,UUAUUU,UUUAAU,UUUACU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | AAUCAU,CUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.