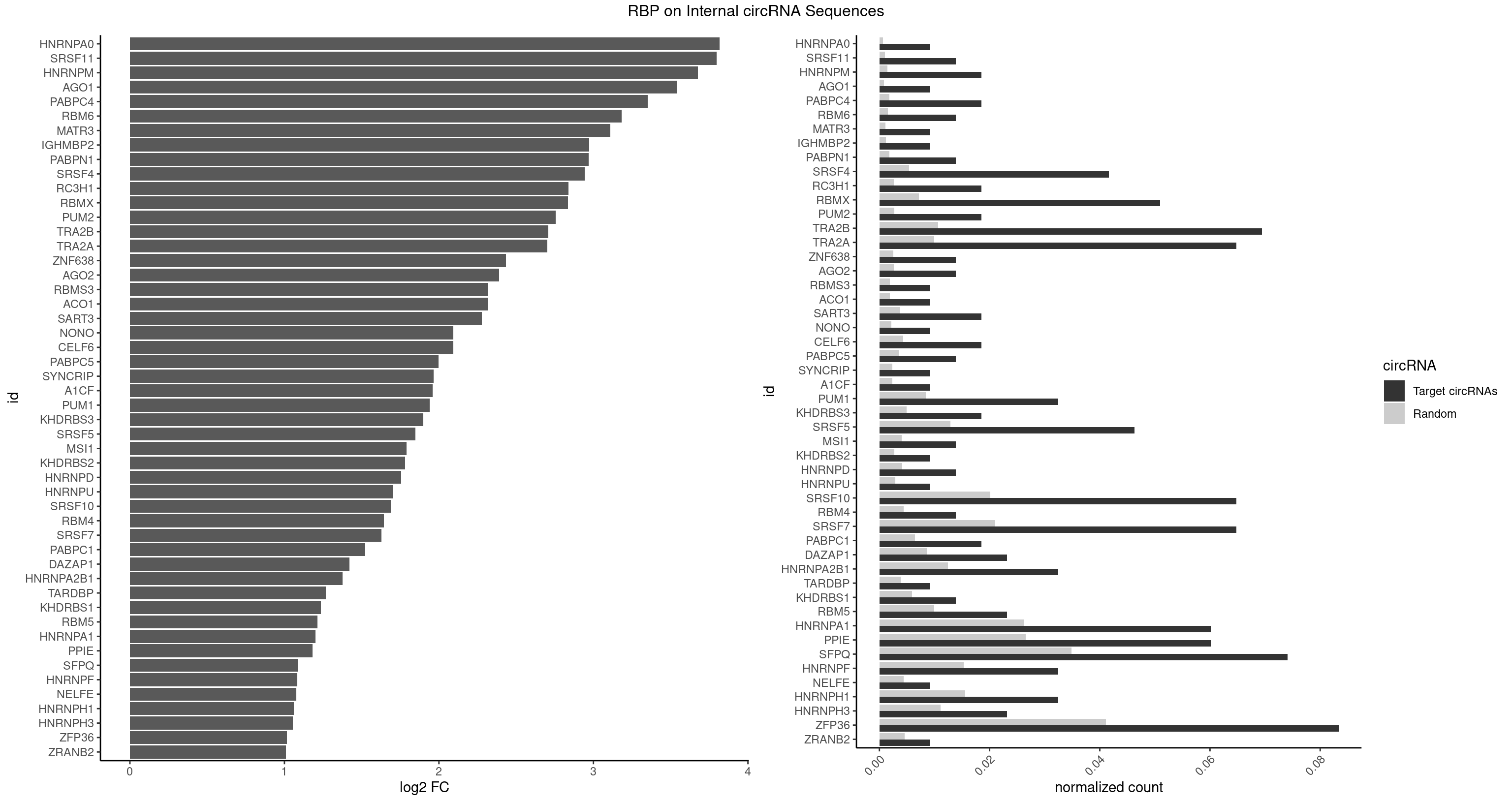
| HNRNPA0 |
1 |
453 |
0.009259259 |
0.0006579386 |
3.814872 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| SRSF11 |
2 |
688 |
0.013888889 |
0.0009985015 |
3.798023 |
AAGAAG |
AAGAAG |
| HNRNPM |
3 |
999 |
0.018518519 |
0.0014492040 |
3.675636 |
AAGGAA,GAAGGA |
AAGGAA,GAAGGA,GGGGGG |
| AGO1 |
1 |
548 |
0.009259259 |
0.0007956130 |
3.540758 |
UGAGGU |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPC4 |
3 |
1251 |
0.018518519 |
0.0018144033 |
3.351402 |
AAAAAA,AAAAAG |
AAAAAA,AAAAAG |
| RBM6 |
2 |
1054 |
0.013888889 |
0.0015289102 |
3.183356 |
AUCCAA,CAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 |
1 |
739 |
0.009259259 |
0.0010724109 |
3.110039 |
AUCUUG |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IGHMBP2 |
1 |
813 |
0.009259259 |
0.0011796520 |
2.972535 |
AAAAAA |
AAAAAA |
| PABPN1 |
2 |
1222 |
0.013888889 |
0.0017723764 |
2.970174 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| SRSF4 |
8 |
3740 |
0.041666667 |
0.0054214720 |
2.942137 |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RC3H1 |
3 |
1786 |
0.018518519 |
0.0025897275 |
2.838097 |
CCUUCU,CUUCUG,UCUGUG |
CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBMX |
10 |
4925 |
0.050925926 |
0.0071387787 |
2.834651 |
AAGAAG,AAGGAA,AAGUGU,AGAAGG,AGGAAG,GAAGGA |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PUM2 |
3 |
1890 |
0.018518519 |
0.0027404447 |
2.756487 |
GUAAAU,UAAAUA,UGUAAA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TRA2B |
14 |
7329 |
0.069444444 |
0.0106226650 |
2.708714 |
AAAGAA,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGAAGG,AGGAAG,GAAGAA,GAAGGA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| TRA2A |
13 |
6871 |
0.064814815 |
0.0099589296 |
2.702261 |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAAGAG |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ZNF638 |
2 |
1773 |
0.013888889 |
0.0025708878 |
2.433593 |
GGUUCU,GUUCUU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| AGO2 |
2 |
1830 |
0.013888889 |
0.0026534924 |
2.387967 |
AAAAAA,AAAGUG |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ACO1 |
1 |
1283 |
0.009259259 |
0.0018607779 |
2.314991 |
CAGUGG |
CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 |
1 |
1283 |
0.009259259 |
0.0018607779 |
2.314991 |
AAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SART3 |
3 |
2634 |
0.018518519 |
0.0038186524 |
2.277833 |
AAAAAA,GAAAAA |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| CELF6 |
3 |
2997 |
0.018518519 |
0.0043447134 |
2.091636 |
GUGAGG,UGUGAG,UGUGGG |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| NONO |
1 |
1498 |
0.009259259 |
0.0021723567 |
2.091636 |
AGAGGA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 |
2 |
2400 |
0.013888889 |
0.0034795387 |
1.996963 |
AGAAAU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SYNCRIP |
1 |
1634 |
0.009259259 |
0.0023694485 |
1.966346 |
AAAAAA |
AAAAAA,UUUUUU |
| A1CF |
1 |
1642 |
0.009259259 |
0.0023810421 |
1.959304 |
GAUCAG |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PUM1 |
6 |
5823 |
0.032407407 |
0.0084401638 |
1.940981 |
AAUGUU,GUAAAU,UAAAUA,UGUAAA,UGUCCA,UUAAUG |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| KHDRBS3 |
3 |
3429 |
0.018518519 |
0.0049707696 |
1.897428 |
AUAAAA,AUUAAA,UAAAUA |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SRSF5 |
9 |
8869 |
0.046296296 |
0.0128544391 |
1.848630 |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GGAAGA,UGCAUC |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| MSI1 |
2 |
2770 |
0.013888889 |
0.0040157442 |
1.790192 |
AGGAAG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| KHDRBS2 |
1 |
1858 |
0.009259259 |
0.0026940701 |
1.781109 |
AUAAAA |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPD |
2 |
2837 |
0.013888889 |
0.0041128408 |
1.755724 |
AAAAAA,AAUUUA |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPU |
1 |
1965 |
0.009259259 |
0.0028491350 |
1.700373 |
AAAAAA |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SRSF10 |
13 |
13860 |
0.064814815 |
0.0200874160 |
1.690032 |
AAAAGA,AAAGAA,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGAGG,AAGGAA,AGAGGA,GAGAGA |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| RBM4 |
2 |
3065 |
0.013888889 |
0.0044432593 |
1.644241 |
CCUUCU,UUCUUG |
CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| SRSF7 |
13 |
14481 |
0.064814815 |
0.0209873716 |
1.626802 |
AAAGGA,AAGAAG,AGAAGA,AGAGAU,AGAGGA,AGAUCA,AGGAAG,GAAGAA,GAGAGA,GAGAUC |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| PABPC1 |
3 |
4443 |
0.018518519 |
0.0064402624 |
1.523777 |
AAAAAA,GAAAAA |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| DAZAP1 |
4 |
5964 |
0.023148148 |
0.0086445016 |
1.421042 |
AAAAAA,AAUUUA,AGGAAG |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPA2B1 |
6 |
8607 |
0.032407407 |
0.0124747476 |
1.377313 |
AAGAAG,AAGGAA,AAUUUA,GAAGGA |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| TARDBP |
1 |
2654 |
0.009259259 |
0.0038476365 |
1.266924 |
GAAUGU |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| KHDRBS1 |
2 |
4064 |
0.013888889 |
0.0058910141 |
1.237343 |
AUAAAA,UAAAAA |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM5 |
4 |
6879 |
0.023148148 |
0.0099705232 |
1.215156 |
AAAAAA,AAGGAA,GAAGGA |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPA1 |
12 |
18070 |
0.060185185 |
0.0261885646 |
1.200471 |
AAAAAA,AAAGAG,AAGAGG,AAUUUA,AGAGGA,AGGAAG,AGUGAA,UAGUGA,UUUAGA |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| PPIE |
12 |
18315 |
0.060185185 |
0.0265436196 |
1.181043 |
AAAAAA,AAAAAU,AAAAUU,AAAUUU,AAUAUA,AAUUUA,AUAAAA,AUAUAA,AUUAAA,UAAAAA,UAAAUA,UAUAAA |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| SFPQ |
15 |
24050 |
0.074074074 |
0.0348548043 |
1.087611 |
AAGAGC,AAGAGG,AAGGAA,AGAGGA,GAAGAA,GAAGAG,GAAGGA,GGAAGA,GGAGAG,UGAUUU,UGGAGA,UUAAUG,UUGAUU |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| HNRNPF |
6 |
10561 |
0.032407407 |
0.0153064921 |
1.082180 |
AAUGUG,AGGAAG,AUGUGG,GAAUGU,UGUGGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| NELFE |
1 |
3028 |
0.009259259 |
0.0043896388 |
1.076795 |
UCUGGC |
CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| HNRNPH1 |
6 |
10717 |
0.032407407 |
0.0155325680 |
1.061027 |
AAUGUG,AGGAAG,AUGUGG,GAAUGU,UGUGGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH3 |
4 |
7688 |
0.023148148 |
0.0111429292 |
1.054768 |
AAUGUG,AUGUGG,GAAUGU,UGUGGG |
AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| ZFP36 |
17 |
28400 |
0.083333333 |
0.0411588414 |
1.017691 |
AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGG,AAAGAA,AAAGGA,AAGAAA,AAGGAA,CCUUCU,GCAAAG,UAAAGA,UAAAUA |
AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| ZRANB2 |
1 |
3173 |
0.009259259 |
0.0045997733 |
1.009334 |
GAGGUU |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |