circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000186298:-:12:110730528:110731901
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000186298:-:12:110730528:110731901 | ENSG00000186298 | ENST00000335007 | - | 12 | 110730529 | 110731901 | 363 | UGAGAGGGUCCAAGCCUGGUAAGAAUGUCCAGCUUCAGGAGAAUGAAAUCAGAGGACUGUGCUUAAAGUCUCGUGAAAUCUUUCUCAGUCAGCCUAUCCUACUAGAACUUGAAGCACCACUCAAAAUAUGUGGUGACAUCCAUGGACAAUACUAUGAUUUGCUGCGACUUUUUGAGUACGGUGGUUUCCCACCAGAAAGCAACUACCUGUUUCUUGGGGACUAUGUGGACAGGGGAAAGCAGUCAUUGGAGACGAUCUGCCUCUUACUGGCCUACAAAAUAAAAUAUCCUGAGAAUUUUUUUCUUCUCAGAGGGAACCAUGAAUGUGCCAGCAUCAACAGAAUUUAUGGAUUUUAUGAUGAAUUGAGAGGGUCCAAGCCUGGUAAGAAUGUCCAGCUUCAGGAGAAUGAAAUC | circ |
| ENSG00000186298:-:12:110730528:110731901 | ENSG00000186298 | ENST00000335007 | - | 12 | 110730529 | 110731901 | 22 | UUAUGAUGAAUUGAGAGGGUCC | bsj |
| ENSG00000186298:-:12:110730528:110731901 | ENSG00000186298 | ENST00000335007 | - | 12 | 110731892 | 110732101 | 210 | CUUGGUAUCGAAGAUGCAAUUAAGUCUUGUCUAAGAUUUUAAGAUUCUAAUUAGACAUUAAAACCAUGUUUAUUAUAAGUGUCCUGAAAAUGAAGCCACUCAGAAAGCUAGUUAAACCAUGUUUUUGCCCCUUUAUCUAAAUCUCGUAUUGUAUUUUGUAUUUUGGCUUCAUUGGACUAAUUGCGAUUUUUCUCUUGCAGUGAGAGGGUC | ie_up |
| ENSG00000186298:-:12:110730528:110731901 | ENSG00000186298 | ENST00000335007 | - | 12 | 110730329 | 110730538 | 210 | UAUGAUGAAUGUAAGUACUUUUCUAUAUCUAAGGAAGAGUUAUUGAAAUUAAGAUAUUUAUCAUAAACAAUUAUCACAUACUUCUGUUAGGAAUUGUGAGAGUUUAGAUGCAGUGGUAUCAUCUCUCAUGUUUUCUAUUUCAUCAACUUGUUUUUAGCUUAGCUUUUUUAGGGGGGUGGGGGAACGGAGUCUCACUCUGUUGCCCAGGCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
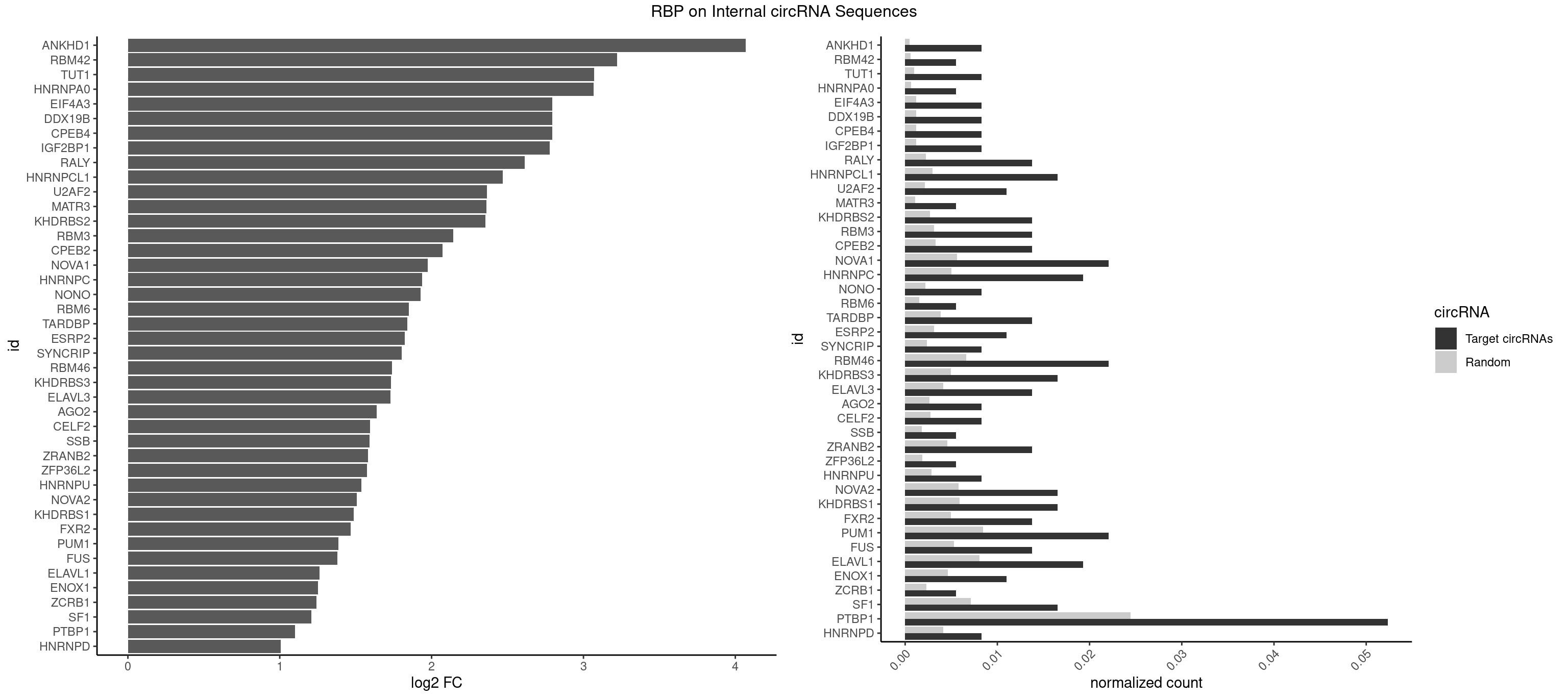
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.008264463 | 0.0004927293 | 4.068054 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM42 | 1 | 407 | 0.005509642 | 0.0005912752 | 3.220057 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| TUT1 | 2 | 678 | 0.008264463 | 0.0009840095 | 3.070177 | AAUACU,CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPA0 | 1 | 453 | 0.005509642 | 0.0006579386 | 3.065934 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| CPEB4 | 2 | 821 | 0.008264463 | 0.0011912456 | 2.794450 | UUUUUU | UUUUUU |
| DDX19B | 2 | 821 | 0.008264463 | 0.0011912456 | 2.794450 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 821 | 0.008264463 | 0.0011912456 | 2.794450 | UUUUUU | UUUUUU |
| IGF2BP1 | 2 | 831 | 0.008264463 | 0.0012057377 | 2.777005 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| RALY | 4 | 1553 | 0.013774105 | 0.0022520629 | 2.612639 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 2062 | 0.016528926 | 0.0029897078 | 2.466917 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| U2AF2 | 3 | 1477 | 0.011019284 | 0.0021419234 | 2.363052 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| MATR3 | 1 | 739 | 0.005509642 | 0.0010724109 | 2.361101 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| KHDRBS2 | 4 | 1858 | 0.013774105 | 0.0026940701 | 2.354099 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBM3 | 4 | 2152 | 0.013774105 | 0.0031201361 | 2.142278 | AAUACU,AGACGA,AUACUA,GAGACG | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CPEB2 | 4 | 2261 | 0.013774105 | 0.0032780993 | 2.071027 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| NOVA1 | 7 | 3872 | 0.022038567 | 0.0056127669 | 1.973246 | AGCACC,AUCAAC,CAGUCA,UCAGUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPC | 6 | 3471 | 0.019283747 | 0.0050316361 | 1.938286 | AUUUUU,CUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NONO | 2 | 1498 | 0.008264463 | 0.0021723567 | 1.927660 | AGAGGA,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM6 | 1 | 1054 | 0.005509642 | 0.0015289102 | 1.849455 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| TARDBP | 4 | 2654 | 0.013774105 | 0.0038476365 | 1.839914 | GAAUGA,GAAUGU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ESRP2 | 3 | 2150 | 0.011019284 | 0.0031172377 | 1.821690 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SYNCRIP | 2 | 1634 | 0.008264463 | 0.0023694485 | 1.802370 | UUUUUU | AAAAAA,UUUUUU |
| RBM46 | 7 | 4554 | 0.022038567 | 0.0066011240 | 1.739247 | AAUGAA,AUGAAA,AUGAAU,AUGAUG,AUGAUU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| KHDRBS3 | 5 | 3429 | 0.016528926 | 0.0049707696 | 1.733452 | AAAUAA,AAUAAA,AUAAAA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ELAVL3 | 4 | 2867 | 0.013774105 | 0.0041563169 | 1.728581 | AUUUAU,AUUUUA,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| AGO2 | 2 | 1830 | 0.008264463 | 0.0026534924 | 1.639029 | GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CELF2 | 2 | 1886 | 0.008264463 | 0.0027346479 | 1.595566 | UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SSB | 1 | 1260 | 0.005509642 | 0.0018274462 | 1.592130 | CUGUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ZRANB2 | 4 | 3173 | 0.013774105 | 0.0045997733 | 1.582324 | AGGGAA,GGUGGU,GUGGUG,UGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ZFP36L2 | 1 | 1277 | 0.005509642 | 0.0018520827 | 1.572810 | AUUUAU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPU | 2 | 1965 | 0.008264463 | 0.0028491350 | 1.536397 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA2 | 5 | 4013 | 0.016528926 | 0.0058171047 | 1.506620 | AGCACC,AGUCAU,AUCAAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 5 | 4064 | 0.016528926 | 0.0058910141 | 1.488405 | AUAAAA,CAAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| FXR2 | 4 | 3434 | 0.013774105 | 0.0049780156 | 1.468316 | AGACGA,GACAGG,GGACAA,GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| PUM1 | 7 | 5823 | 0.022038567 | 0.0084401638 | 1.384688 | AGAAUU,CAGAAU,CCAGAA,GAAUUG,GUCCAG,UGUCCA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| FUS | 4 | 3648 | 0.013774105 | 0.0052881452 | 1.381125 | CGGUGG,UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL1 | 6 | 5554 | 0.019283747 | 0.0080503280 | 1.260266 | AUUUAU,UGAUUU,UGGUUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ENOX1 | 3 | 3195 | 0.011019284 | 0.0046316558 | 1.250430 | GGACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZCRB1 | 1 | 1605 | 0.005509642 | 0.0023274215 | 1.243226 | GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SF1 | 5 | 4931 | 0.016528926 | 0.0071474739 | 1.209488 | AUACUA,CAGUCA,UACUAG,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PTBP1 | 18 | 16854 | 0.052341598 | 0.0244263326 | 1.099521 | ACUUUU,AUUUUU,CCUCUU,CUCUUA,CUUCUC,CUUUCU,CUUUUU,UCUUCU,UCUUUC,UUCUUC,UUCUUG,UUUCCC,UUUCUU,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| HNRNPD | 2 | 2837 | 0.008264463 | 0.0041128408 | 1.006786 | AAUUUA,AUUUAU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
RBP on BSJ (Exon Only)
Plot
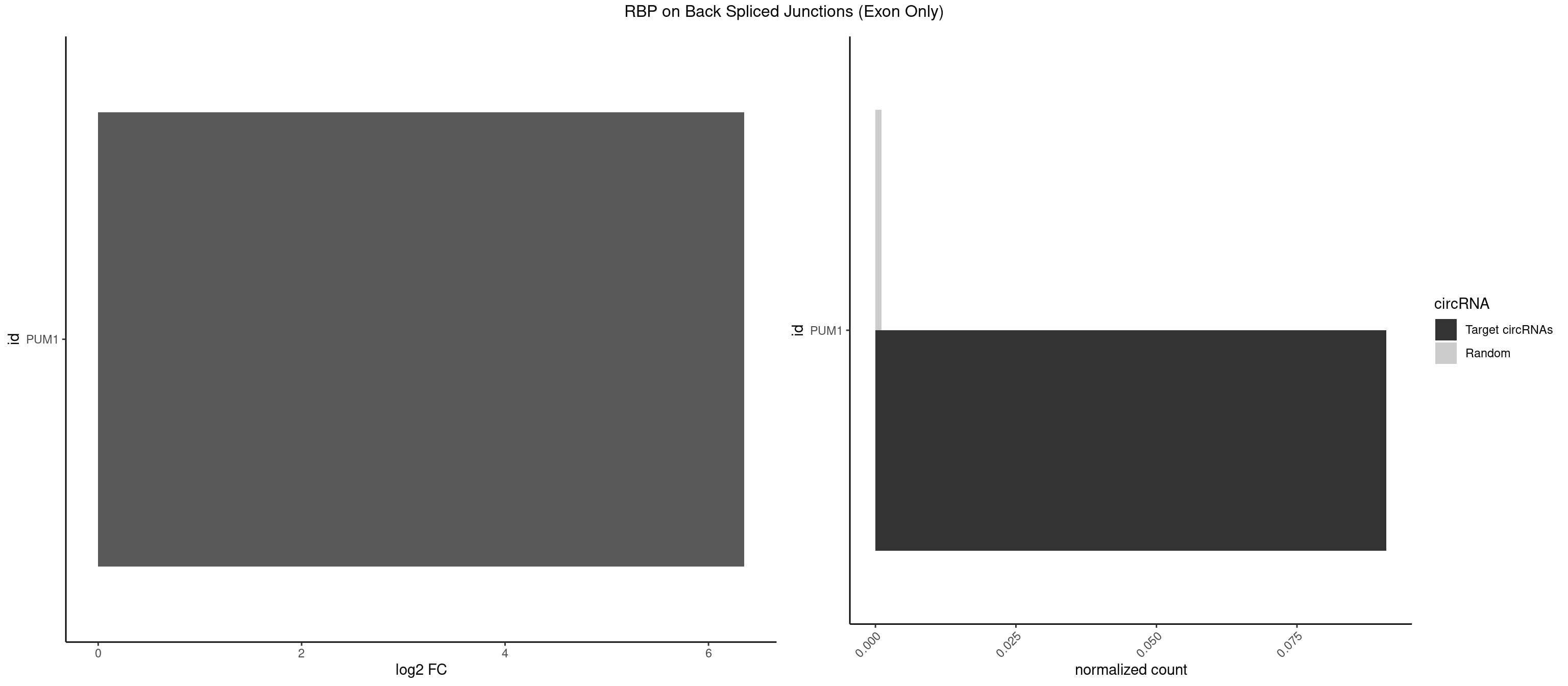
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM1 | 1 | 16 | 0.09090909 | 0.00111344 | 6.351329 | GAAUUG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
RBP on BSJ (Exon and Intron)
Plot
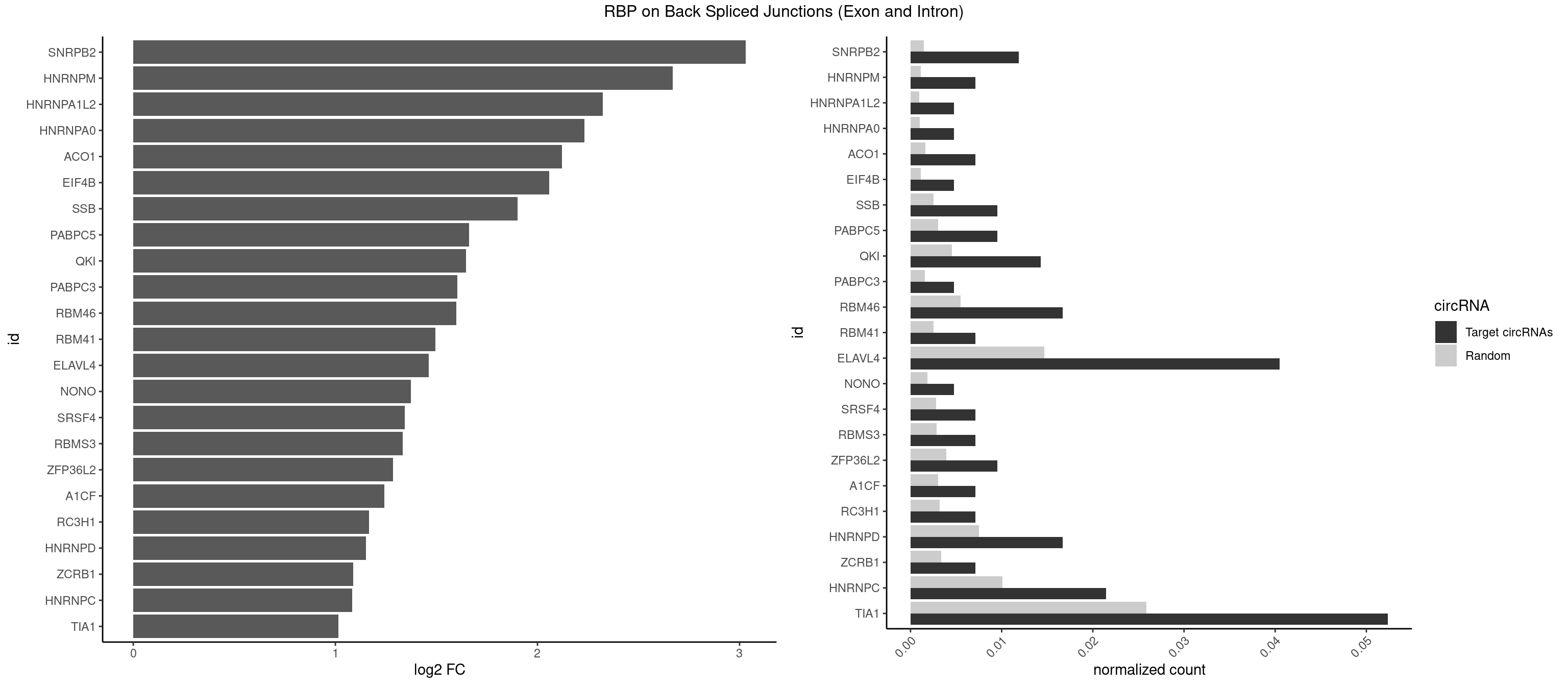
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPB2 | 4 | 288 | 0.011904762 | 0.001456066 | 3.031391 | GUAUUG,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPM | 2 | 222 | 0.007142857 | 0.001123539 | 2.668451 | AAGGAA,GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.000952237 | 2.322146 | UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.001012696 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.001642483 | 2.120623 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.001143692 | 2.057840 | UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SSB | 3 | 505 | 0.009523810 | 0.002549375 | 1.901395 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.003007860 | 1.662801 | AGAAAG,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| QKI | 5 | 904 | 0.014285714 | 0.004559653 | 1.647577 | ACUAAU,AUCAUA,AUCUAA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.001566909 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RBM46 | 6 | 1091 | 0.016666667 | 0.005501814 | 1.598986 | AAUGAA,AUCAUA,AUGAAG,AUGAAU,AUGAUG,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM41 | 2 | 502 | 0.007142857 | 0.002534260 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ELAVL4 | 16 | 2916 | 0.040476190 | 0.014696695 | 1.461582 | AUCUAA,AUUUAU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UUGUAU,UUUAUU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| NONO | 1 | 364 | 0.004761905 | 0.001838976 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SRSF4 | 2 | 558 | 0.007142857 | 0.002816405 | 1.342647 | AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBMS3 | 2 | 562 | 0.007142857 | 0.002836558 | 1.332360 | CUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.003904676 | 1.286336 | AUUUAU,UAUUUA,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.003017936 | 1.242939 | UAAUUA,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.003184200 | 1.165570 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.007502015 | 1.151615 | AGAUAU,AUUUAU,UAUUUA,UUAGGA,UUAGGG,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | AAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.010111850 | 1.083489 | AUUUUU,CUUUUU,GGGGGG,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.025901854 | 1.015987 | AUUUUG,AUUUUU,CUUUUC,CUUUUU,GUUUUC,GUUUUU,UAUUUU,UUUAUU,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.