circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000139746:-:13:79318820:79320710
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000139746:-:13:79318820:79320710 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79318821 | 79320710 | 1235 | UUUCAGGAAGAGUCUUUGGUGGAUGACUCAUUACUUCAAGAUGAUGAUGAAGAAGAAGAGGACAAUGAAUCUCGUUCUUGGAGAAGAUGAUUUGACUGAUCAUUGAUCUGCAUAUGCUAGAACUCUACCUGUGUUUCAUUAGUAUUAUCUAAUGUACUUUUACAUAUUUGUAAAAACAAUUUUUGGUAAAAUGUGAUGAAGAUGGAUUUCACAAAUAGACAAAAAAGAAGAAAACUACCUUCUGAUCUUGUAUUUUGAAAGAUUGAUGUUUGCAUUUUACUUCAGUAAACAAUUGCUAAAGACAUCACACUAGAAACAUAUGCAAUGUUUUUAUUACAUACUUCUACUGGACAUCACAGAAUUCUUUGGGUUCUUUGUAAUUUAAUGAAUAGGUCUGAAAACUUAUGACCAAUACUUGUUAUAACUUAGAGGACUUUGUUUUAUUCCAAAUAAGGAAUGAAUUUGCAUUUAAAAUCUUAAUGAAUGUUUUCAAAACUGAAUAGAUAACAUAGUACUCUAACUAAAGUCUCCAAGUUAUGUAUUAUAAUAUUACAUAGUAGUAUGCUUAGGCUUUACUAUGUAUUAGCCUUUUGUUGGACUGUGUAUGUAUUUUACCAUAUGGGUUUUAAUGAUAAUGGUGUAUGACUGCUUUACAUGAGUCCUUAUGCAUCCAGAUGUUAUAAUAAAGUGGAAUGGUCUCUUUAAAAAAAAAAAAGGAAAGAAAAGAGAAAAGCAAUGACAAAAAAAAAAAAAAAAAAAAAGCCAAGACAAUGUUCCUUGAUUUAAAAAAAAAAAGAAAAGAAAAAAGAAAAGAAAAAAAUUAAAAUAGACCAUUCCAGAUGGUGAUCUACCCUUCCCCCCAAUUAUCACAAAAGGAGCUAUAAUCACUAACUUAUUCUUAAUAAUAAUGGUUACUGGUACCCUUACAAUAAUGUACAAUCCAAUGUUUAAAAAUUAUACCACUUGAGUUUGGUUUGAUCCUAUAAAUUUUCAACAAAUGUCUUAAAAUUUAAAAGCAGGCUGAUUAGUUUGGAACCAGACUGCAAGUAGAGCUAACAUUUGGUGAAUAAGAAAACAUCACAUUACAUUUUGGAUGUAUGAAAGAAAACUUGACCAUUUGAAGAUAUAUGAAUAAAGAAAUGUACUAUAGAUACUACUUUAGGAAAACACUGUUUGUAGUCGUGAAAGGUGCCAAAUUGGCAGAGGCUCACGUUUCUUCUCUUUACACCAAACAGUUUCAGGAAGAGUCUUUGGUGGAUGACUCAUUACUUCAAGAUGAUGAUGA | circ |
| ENSG00000139746:-:13:79318820:79320710 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79318821 | 79320710 | 22 | ACACCAAACAGUUUCAGGAAGA | bsj |
| ENSG00000139746:-:13:79318820:79320710 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79320701 | 79320910 | 210 | UGCAAGCAAUAGAAUAGAUCAUAUAGAGAAACCAUUGAGUGGUUAAAUGUUGCUUUUCUUUGUUUUGUGAAUGUUCUAGCUAUAAAAAUACCUUUUGUAUUCAACUCUGUUGAGAGAGAUGAGUAUUUAAAAGUGUCACCUUUUUUUUCUUUUCUUUUUUUUUUUUUUGGGUAAAUUCCUAAAUACAUUUUAACCUAAAGUUUCAGGAAG | ie_up |
| ENSG00000139746:-:13:79318820:79320710 | ENSG00000139746 | MSTRG.8946.3 | - | 13 | 79318621 | 79318830 | 210 | CACCAAACAGGUAUUACAAAGAAAGUAAAUCUUUCAAAGGCCAGUUAAAUCUCCAUGCUGAUUUUUGGCUUUAUAAUUUGAUUAGUAUGUUUUAGCCUGAUAUUUCAAAAAUUAUAAAAUUCCCUUACAAUGUAACUGAAAAUACAUAUUCUGUUUAUUUCAGUCCAUGAAUAGCUAAUCAGUUUUCCUAGAACUUAAAAUGUUUUAAGC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
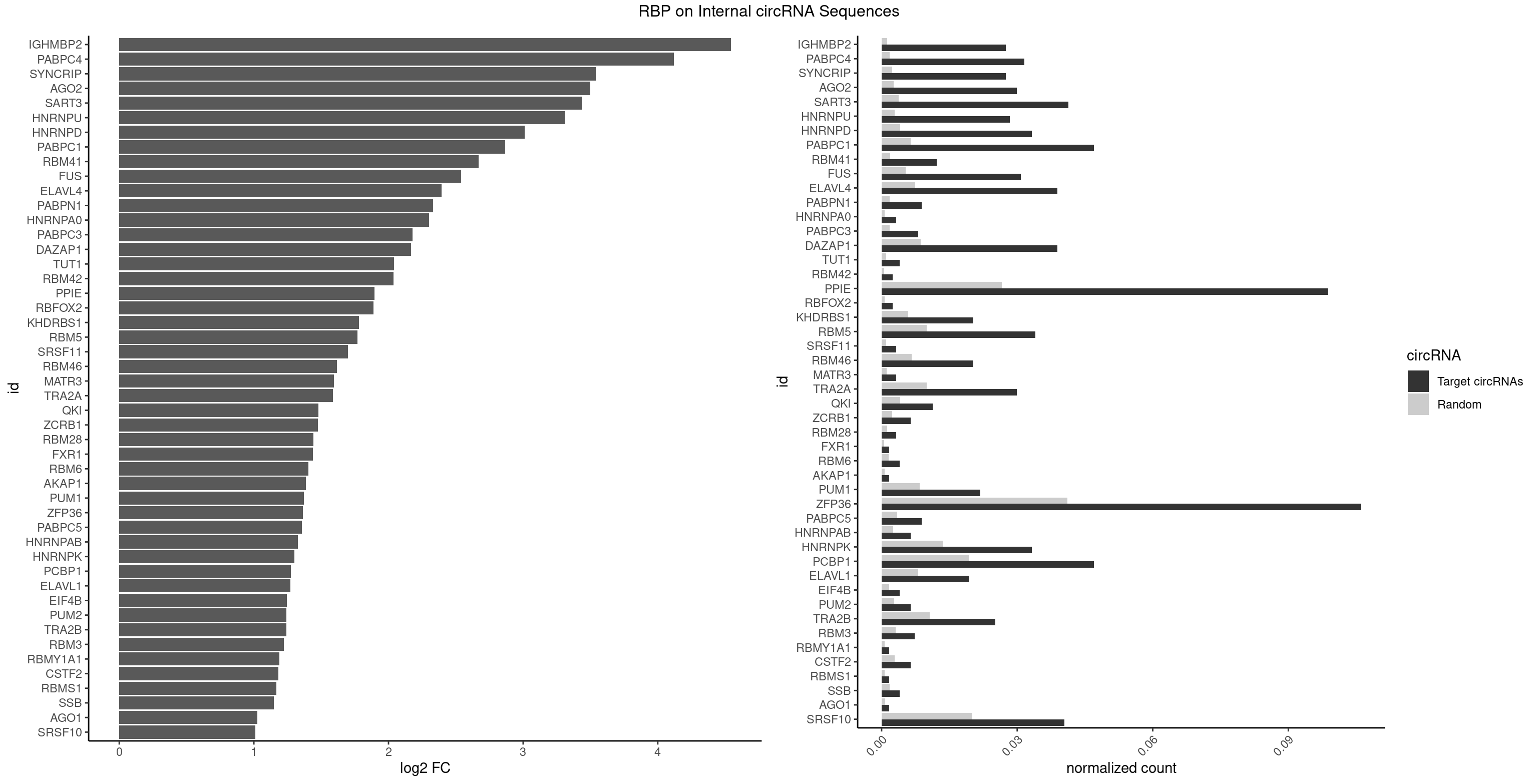
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 33 | 813 | 0.027530364 | 0.0011796520 | 4.544590 | AAAAAA | AAAAAA |
| PABPC4 | 38 | 1251 | 0.031578947 | 0.0018144033 | 4.121396 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| SYNCRIP | 33 | 1634 | 0.027530364 | 0.0023694485 | 3.538401 | AAAAAA | AAAAAA,UUUUUU |
| AGO2 | 36 | 1830 | 0.029959514 | 0.0026534924 | 3.497050 | AAAAAA,AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SART3 | 50 | 2634 | 0.041295547 | 0.0038186524 | 3.434851 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPU | 34 | 1965 | 0.028340081 | 0.0028491350 | 3.314248 | AAAAAA,CCCCCC | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| HNRNPD | 40 | 2837 | 0.033198381 | 0.0041128408 | 3.012906 | AAAAAA,AAUUUA,AGAUAU,UUAGAG,UUAGGA,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| PABPC1 | 57 | 4443 | 0.046963563 | 0.0064402624 | 2.866350 | AAAAAA,AAAAAC,ACAAAU,ACUAAC,AGAAAA,CAAACA,CAAAUA,CUAACA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM41 | 14 | 1318 | 0.012145749 | 0.0019115000 | 2.667674 | AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| FUS | 37 | 3648 | 0.030769231 | 0.0052881452 | 2.540655 | AAAAAA,UGGUGA,UGGUGG,UGGUGU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL4 | 47 | 5105 | 0.038866397 | 0.0073996354 | 2.392997 | AAAAAA,AUCUAA,UAUCUA,UAUUUU,UCUAAU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| PABPN1 | 10 | 1222 | 0.008906883 | 0.0017723764 | 2.329236 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| HNRNPA0 | 3 | 453 | 0.003238866 | 0.0006579386 | 2.299464 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| PABPC3 | 9 | 1234 | 0.008097166 | 0.0017897669 | 2.177645 | AAAAAC,AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| DAZAP1 | 47 | 5964 | 0.038866397 | 0.0086445016 | 2.168669 | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGUAAA,AGUAUG,AGUUUG,UAGGAA,UAGUAU,UAGUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| TUT1 | 4 | 678 | 0.004048583 | 0.0009840095 | 2.040673 | AAUACU,AGAUAC,CAAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM42 | 2 | 407 | 0.002429150 | 0.0005912752 | 2.038550 | AACUAA,AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| PPIE | 121 | 18315 | 0.098785425 | 0.0265436196 | 1.895933 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAUA,AUUUAA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUU,UAAUAA,UAAUAU,UAAUUU,UAUAAA,UAUAAU,UAUUAU,UAUUUU,UUAAAA,UUAAUA,UUAUAA,UUAUUA,UUUAAA,UUUAAU,UUUAUU,UUUUAA,UUUUAU,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| RBFOX2 | 2 | 452 | 0.002429150 | 0.0006564894 | 1.887608 | UGACUG | UGACUG,UGCAUG |
| KHDRBS1 | 24 | 4064 | 0.020242915 | 0.0058910141 | 1.780829 | AUUUAA,GAAAAC,UAAAAA,UAAAAG,UUAAAA,UUUUAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM5 | 41 | 6879 | 0.034008097 | 0.0099705232 | 1.770137 | AAAAAA,AAGGAA,AAGGAG,AAGGUG,CCCCCC,CUUCUC,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| SRSF11 | 3 | 688 | 0.003238866 | 0.0009985015 | 1.697652 | AAGAAG | AAGAAG |
| RBM46 | 24 | 4554 | 0.020242915 | 0.0066011240 | 1.616633 | AAUGAA,AAUGAU,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| MATR3 | 3 | 739 | 0.003238866 | 0.0010724109 | 1.594631 | AAUCUU,AUCUUA,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| TRA2A | 36 | 6871 | 0.029959514 | 0.0099589296 | 1.588952 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| QKI | 13 | 2805 | 0.011336032 | 0.0040664663 | 1.479068 | ACACUA,ACUAAC,ACUCAU,ACUUAU,AUCUAA,AUCUAC,CACACU,CACUAA,CUAACA,UAAUCA,UCUAAU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZCRB1 | 7 | 1605 | 0.006477733 | 0.0023274215 | 1.476756 | AAUUAA,AUUUAA,GAUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM28 | 3 | 822 | 0.003238866 | 0.0011926949 | 1.441264 | AGUAGA,AGUAGU,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FXR1 | 1 | 411 | 0.001619433 | 0.0005970720 | 1.439512 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM6 | 4 | 1054 | 0.004048583 | 0.0015289102 | 1.404913 | AAUCCA,AUCCAA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| AKAP1 | 1 | 426 | 0.001619433 | 0.0006188101 | 1.387920 | AUAUAU | AUAUAU,UAUAUA |
| PUM1 | 26 | 5823 | 0.021862348 | 0.0084401638 | 1.373105 | AAUAUU,AAUGUU,AGAAUU,AGAUAA,CAGAAU,CUUGUA,UAAUAU,UAAUGU,UACAUA,UAGAUA,UGUAAA,UGUAAU,UGUACA,UUAAUG,UUGUAG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ZFP36 | 130 | 28400 | 0.106072874 | 0.0411588414 | 1.365781 | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAUAA,AACAAA,AAGAAA,AAGCAA,AAGGAA,AAUAAA,AAUAAG,ACAAAU,ACCUGU,ACCUUC,ACUUCU,ACUUGU,ACUUUU,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGG,AUUUAA,CAAACA,CAAAUA,CAAGUA,CCCUUC,CCUUCC,CCUUCU,CCUUUU,GGAAAG,UAAACA,UAAAGA,UAAGGA,UCUUCU,UCUUGU,UUUAAA,UUUAAU,UUUAUU | AAAAAA,AAAAAC,AAAAAG,AAAAAU,AAAACA,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAACAA,AAAGAA,AAAGCA,AAAGGA,AAAGUA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAA,AAUAAG,ACAAAC,ACAAAG,ACAAAU,ACAAGC,ACAAGG,ACAAGU,ACCUCC,ACCUCU,ACCUGC,ACCUGU,ACCUUC,ACCUUU,ACUUCC,ACUUCU,ACUUGC,ACUUGU,ACUUUC,ACUUUU,AGAAAG,AGCAAA,AGGAAA,AGUAAA,AUAAAC,AUAAAG,AUAAAU,AUAAGC,AUAAGG,AUAAGU,AUUUAA,AUUUAU,CAAACA,CAAAGA,CAAAUA,CAAGCA,CAAGGA,CAAGUA,CCCUCC,CCCUCU,CCCUGC,CCCUGU,CCCUUC,CCCUUU,CCUUCC,CCUUCU,CCUUGC,CCUUGU,CCUUUC,CCUUUU,GCAAAG,GGAAAG,GUAAAG,UAAACA,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UAAGUA,UAUUUA,UCCUCC,UCCUCU,UCCUGC,UCCUGU,UCCUUC,UCCUUU,UCUUCC,UCUUCU,UCUUGC,UCUUGU,UCUUUC,UCUUUU,UUAUUU,UUGUGC,UUGUGG,UUUAAA,UUUAAU,UUUAUA,UUUAUU |
| PABPC5 | 10 | 2400 | 0.008906883 | 0.0034795387 | 1.356025 | AGAAAA,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPAB | 7 | 1782 | 0.006477733 | 0.0025839306 | 1.325922 | AAAGAC,AAGACA,AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPK | 40 | 9304 | 0.033198381 | 0.0134848428 | 1.299774 | AAAAAA,ACCCUU,CCAAAC,CCCCAA,CCCCCA,CCCCCC,UCCCCC | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| PCBP1 | 57 | 13384 | 0.046963563 | 0.0193975949 | 1.275664 | AAAAAA,AAAUUA,AAUUAA,ACAUUA,ACCAAA,AUUAAA,AUUCCA,CAAUCC,CAAUUA,CAUUCC,CCAAAC,CCAUUC,CCCCAA,CCCCCC,CCCUUA,CCCUUC,CCUUAC,CCUUCC,CUACCC,CUUAAA,CUUCCC,UUAGAG,UUAGGA | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| ELAVL1 | 23 | 5554 | 0.019433198 | 0.0080503280 | 1.271404 | CCCCCC,UAGUUU,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUGAUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| EIF4B | 4 | 1179 | 0.004048583 | 0.0017100607 | 1.243370 | CUUGGA,GUUGGA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PUM2 | 7 | 1890 | 0.006477733 | 0.0027404447 | 1.241079 | UACAUA,UAGAUA,UGUAAA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TRA2B | 30 | 7329 | 0.025101215 | 0.0106226650 | 1.240611 | AAAGAA,AAGAAG,AAGGAA,AAUAAG,AGAAGA,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM3 | 8 | 2152 | 0.007287449 | 0.0031201361 | 1.223805 | AAAACU,AAACUA,AAUACU,AUACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBMY1A1 | 1 | 489 | 0.001619433 | 0.0007101099 | 1.189375 | CAAGAC | ACAAGA,CAAGAC |
| CSTF2 | 7 | 1967 | 0.006477733 | 0.0028520334 | 1.183498 | GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBMS1 | 1 | 497 | 0.001619433 | 0.0007217036 | 1.166011 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SSB | 4 | 1260 | 0.004048583 | 0.0018274462 | 1.147588 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| AGO1 | 1 | 548 | 0.001619433 | 0.0007956130 | 1.025350 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRSF10 | 49 | 13860 | 0.040485830 | 0.0200874160 | 1.011125 | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AGACAA,AGAGAA,AGAGGA,GAAAGA,GACAAA,GAGAAA,GAGAAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon Only)
Plot
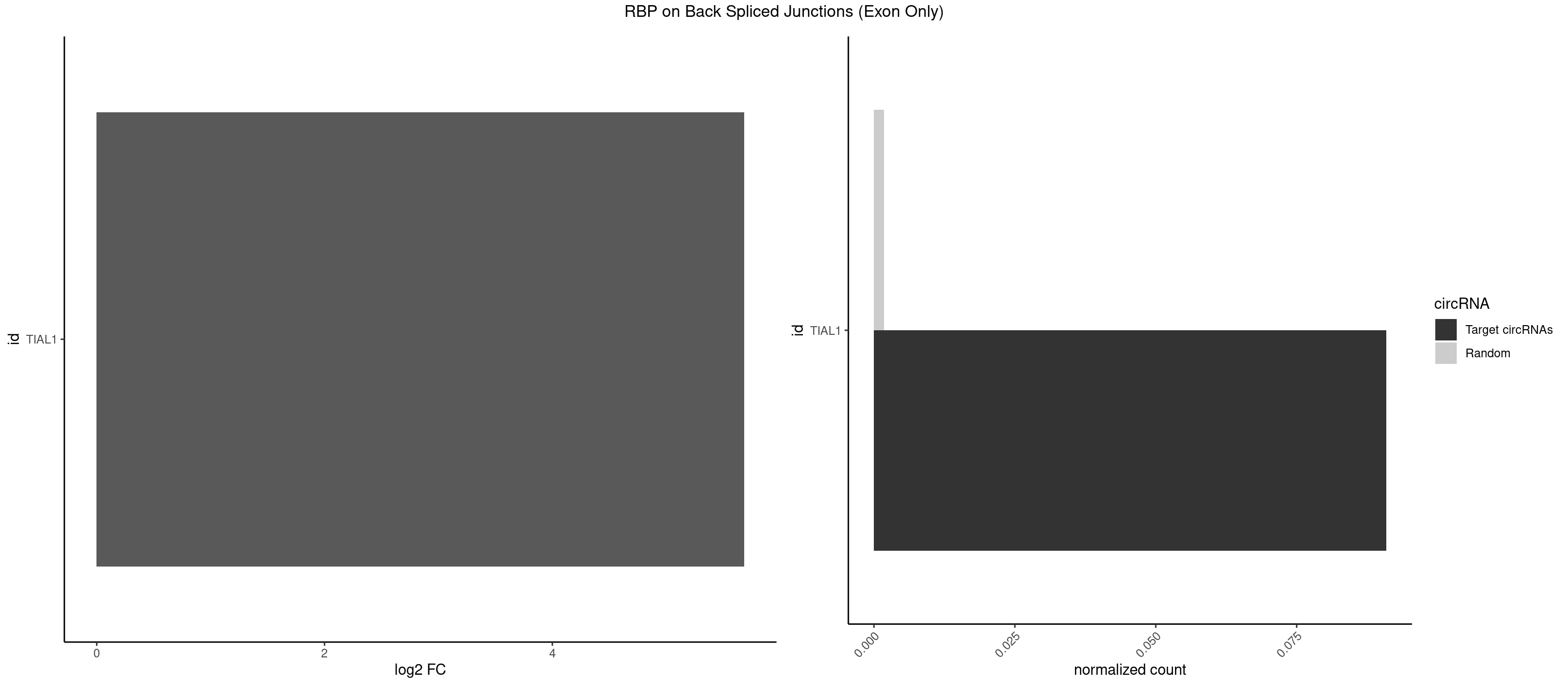
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TIAL1 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | CAGUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
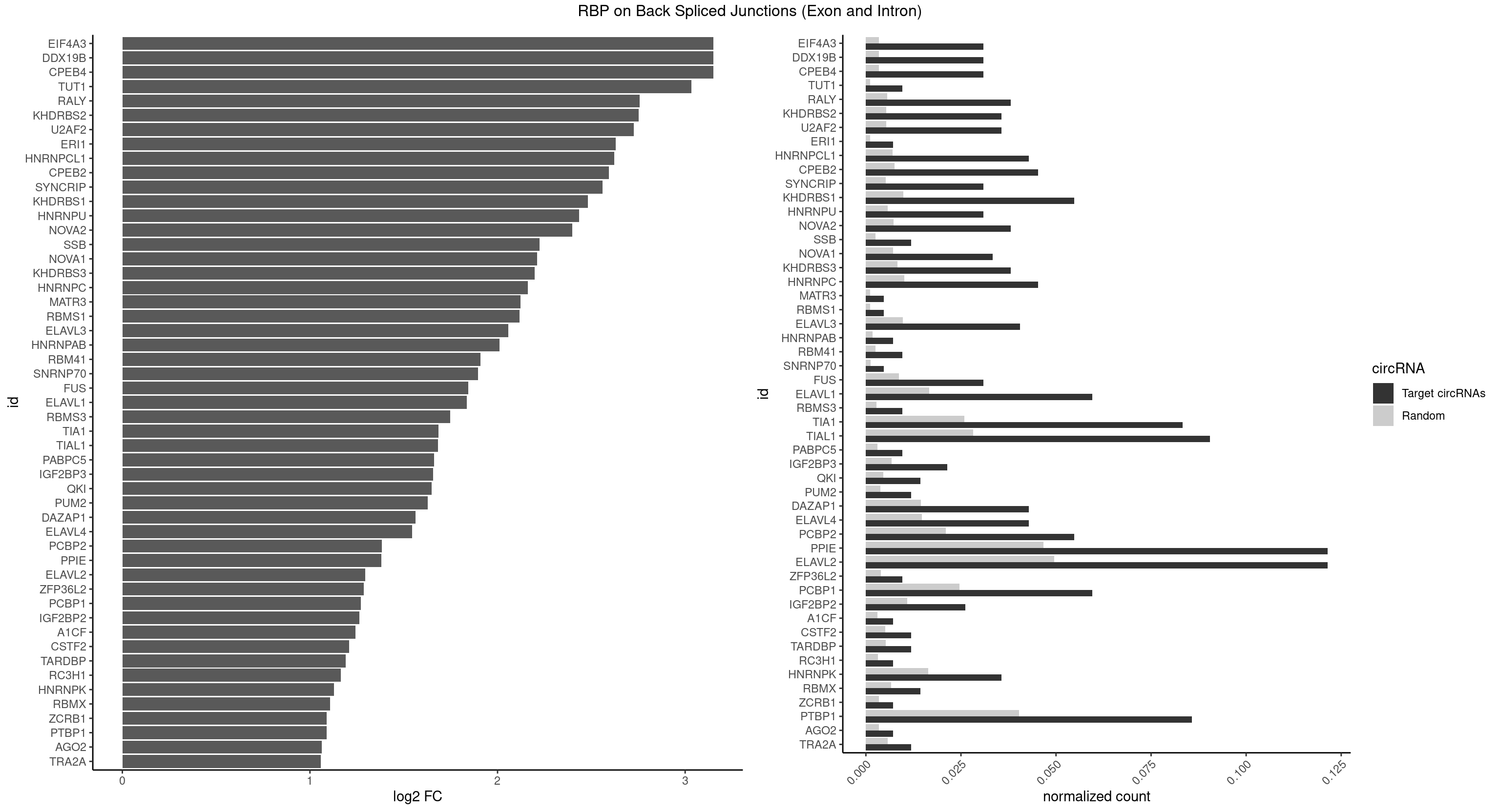
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CPEB4 | 12 | 691 | 0.030952381 | 0.003486497 | 3.150200 | UUUUUU | UUUUUU |
| DDX19B | 12 | 691 | 0.030952381 | 0.003486497 | 3.150200 | UUUUUU | UUUUUU |
| EIF4A3 | 12 | 691 | 0.030952381 | 0.003486497 | 3.150200 | UUUUUU | UUUUUU |
| TUT1 | 3 | 230 | 0.009523810 | 0.001163845 | 3.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RALY | 15 | 1117 | 0.038095238 | 0.005632809 | 2.757684 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| KHDRBS2 | 14 | 1051 | 0.035714286 | 0.005300282 | 2.752360 | AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 | 14 | 1071 | 0.035714286 | 0.005401048 | 2.725190 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.001153769 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPCL1 | 17 | 1381 | 0.042857143 | 0.006962918 | 2.621772 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 18 | 1487 | 0.045238095 | 0.007496977 | 2.593157 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| SYNCRIP | 12 | 1041 | 0.030952381 | 0.005249899 | 2.559689 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS1 | 22 | 1945 | 0.054761905 | 0.009804514 | 2.481655 | AUAAAA,AUUUAA,CUAAAU,UAAAAA,UAAAAG,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPU | 12 | 1136 | 0.030952381 | 0.005728537 | 2.433812 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA2 | 15 | 1434 | 0.038095238 | 0.007229948 | 2.397554 | AGAUCA,CCUAGA,UAGAUC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SSB | 4 | 505 | 0.011904762 | 0.002549375 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| NOVA1 | 13 | 1428 | 0.033333333 | 0.007199718 | 2.210953 | UCAGUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| KHDRBS3 | 15 | 1646 | 0.038095238 | 0.008298065 | 2.198764 | AUAAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPC | 18 | 2006 | 0.045238095 | 0.010111850 | 2.161491 | AUUUUU,CUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.001093309 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.001098347 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ELAVL3 | 16 | 1928 | 0.040476190 | 0.009718863 | 2.058214 | AUUUUA,UAUUUA,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.001773478 | 2.009919 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM41 | 3 | 502 | 0.009523810 | 0.002534260 | 1.909974 | AUACAU,UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| FUS | 12 | 1711 | 0.030952381 | 0.008625554 | 1.843361 | UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ELAVL1 | 24 | 3309 | 0.059523810 | 0.016676743 | 1.835629 | UAUUUA,UGAUUU,UGUUUU,UUAUUU,UUGAUU,UUGUUU,UUUAUU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.002836558 | 1.747397 | AUAUAG,CAUAUA,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TIA1 | 34 | 5140 | 0.083333333 | 0.025901854 | 1.685838 | AUUUUU,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,UUAUUU,UUUAUU,UUUUCU,UUUUGG,UUUUGU,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 | 37 | 5597 | 0.090476190 | 0.028204353 | 1.681620 | AGUUUU,AUUUUA,AUUUUU,CAGUUU,CUUUUC,CUUUUG,CUUUUU,GUUUUC,UAAAUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.003007860 | 1.662801 | AGAAAG,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 8 | 1348 | 0.021428571 | 0.006796655 | 1.656639 | AAAUAC,AAAUUC,AAUACA,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| QKI | 5 | 904 | 0.014285714 | 0.004559653 | 1.647577 | AUCAUA,CUAAUC,UAACCU,UAAUCA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PUM2 | 4 | 764 | 0.011904762 | 0.003854293 | 1.627001 | GUAAAU,UAAAUA,UACAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| DAZAP1 | 17 | 2878 | 0.042857143 | 0.014505240 | 1.562962 | AGGAAG,AGUAAA,AGUAUG,AGUUAA,UAGUAU,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| ELAVL4 | 17 | 2916 | 0.042857143 | 0.014696695 | 1.544044 | UAUUUA,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| PCBP2 | 22 | 4164 | 0.054761905 | 0.020984482 | 1.383850 | AAACCA,AAAUUA,AAAUUC,AAUUCC,ACCAAA,AUUCCC,CCCUUA,CUUAAA,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| PPIE | 50 | 9262 | 0.121428571 | 0.046669690 | 1.379550 | AAAAAU,AAAAUA,AAAAUU,AAAUUA,AAUUAU,AUAAAA,AUAAUU,AUAUUU,AUUAUA,AUUUAA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUUU,UAUAAA,UAUAAU,UAUUUA,UUAAAA,UUAAAU,UUAUAA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL2 | 50 | 9826 | 0.121428571 | 0.049511286 | 1.294279 | AAUUUG,AUAUUU,AUUUAA,AUUUUA,AUUUUU,CAUUUU,CUUUUU,GAUUUU,GUUUUA,GUUUUC,UAAUUU,UAUGUU,UAUUUA,UCUUUU,UGAUUU,UGUAUU,UUAUUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAUA,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.003904676 | 1.286336 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PCBP1 | 24 | 4891 | 0.059523810 | 0.024647320 | 1.272036 | AAACCA,AAAUUA,AAAUUC,AAUUCC,ACCAAA,AUUCCC,CCAAAC,CCCUUA,CCUUAC,CUUAAA,UUUUUU | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| IGF2BP2 | 10 | 2163 | 0.026190476 | 0.010902862 | 1.264335 | AAAUAC,AAAUUC,AAUACA,CAAACA,CAACUC,CCAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| A1CF | 2 | 598 | 0.007142857 | 0.003017936 | 1.242939 | AUAAUU,AUCAGU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.005154172 | 1.207726 | GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.005214631 | 1.190902 | GAAUGU,GUGAAU,GUUUUG,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RC3H1 | 2 | 631 | 0.007142857 | 0.003184200 | 1.165570 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPK | 14 | 3244 | 0.035714286 | 0.016349254 | 1.127276 | CCAAAC,UCCCUU,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| RBMX | 5 | 1316 | 0.014285714 | 0.006635429 | 1.106311 | AAGUAA,AAGUGU,ACCAAA,AGGAAG,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PTBP1 | 35 | 8003 | 0.085714286 | 0.040326481 | 1.087808 | AUUUUU,CCUUUU,CUCCAU,CUUUUC,CUUUUU,UCUUUC,UCUUUU,UUAUUU,UUCUUU,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | ACUUAA,AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO2 | 2 | 677 | 0.007142857 | 0.003415961 | 1.064210 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.005713422 | 1.059112 | AAAGAA,AAGAAA,AGAAAG,AGGAAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.