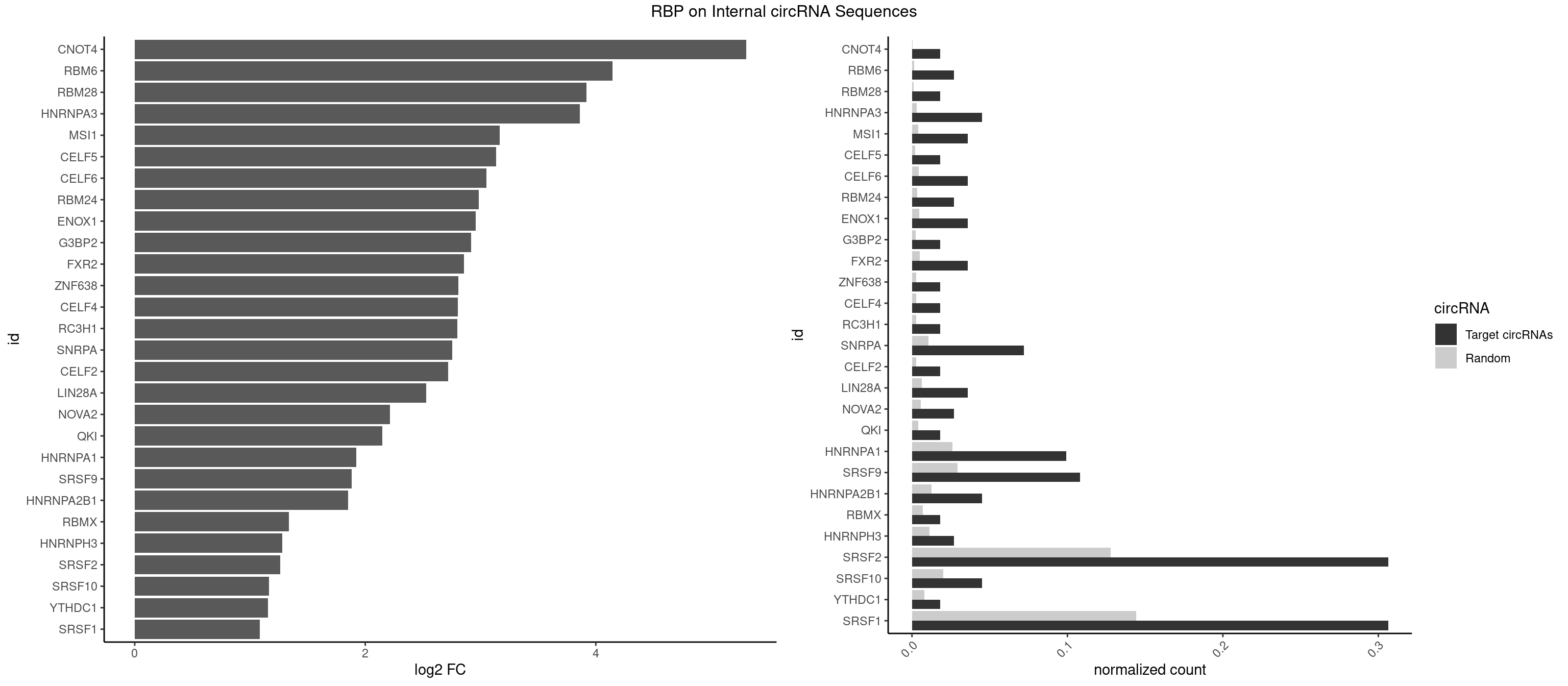
| CNOT4 |
1 |
314 |
0.01801802 |
0.0004564992 |
5.302684 |
GACAGA |
GACAGA |
| RBM6 |
2 |
1054 |
0.02702703 |
0.0015289102 |
4.143827 |
AUCCAG,CAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM28 |
1 |
822 |
0.01801802 |
0.0011926949 |
3.917143 |
AGUAGU |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 |
4 |
2140 |
0.04504505 |
0.0031027457 |
3.859751 |
AGGAGC,GCCAAG,GGAGCC |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| MSI1 |
3 |
2770 |
0.03603604 |
0.0040157442 |
3.165701 |
AGGAGG |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF5 |
1 |
1415 |
0.01801802 |
0.0020520728 |
3.134287 |
GUGUGG |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF6 |
3 |
2997 |
0.03603604 |
0.0043447134 |
3.052107 |
GUGAGG,GUGUGG,UGUGAG |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM24 |
2 |
2357 |
0.02702703 |
0.0034172229 |
2.983507 |
AGUGUG,GUGUGG |
AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ENOX1 |
3 |
3195 |
0.03603604 |
0.0046316558 |
2.959840 |
AGACAG,CAGACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| G3BP2 |
1 |
1644 |
0.01801802 |
0.0023839405 |
2.918020 |
GGAUUG |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| FXR2 |
3 |
3434 |
0.03603604 |
0.0049780156 |
2.855798 |
AGACAG,GACAGA |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ZNF638 |
1 |
1773 |
0.01801802 |
0.0025708878 |
2.809102 |
GUUCGU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF4 |
1 |
1782 |
0.01801802 |
0.0025839306 |
2.801801 |
GUGUGG |
GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RC3H1 |
1 |
1786 |
0.01801802 |
0.0025897275 |
2.798568 |
UCUGUG |
CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SNRPA |
7 |
7380 |
0.07207207 |
0.0106965744 |
2.752291 |
AGCAGU,AGUAGU,CAGUAG,GAGCAG,GCAGUA,GUAGUC |
ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| CELF2 |
1 |
1886 |
0.01801802 |
0.0027346479 |
2.720013 |
GUCUGU |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| LIN28A |
3 |
4315 |
0.03603604 |
0.0062547643 |
2.526413 |
GGAGGA,UGGAGG |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NOVA2 |
2 |
4013 |
0.02702703 |
0.0058171047 |
2.216030 |
AGGCAU,GAGGCA |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| QKI |
1 |
2805 |
0.01801802 |
0.0040664663 |
2.147593 |
CUACUC |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPA1 |
10 |
18070 |
0.09909910 |
0.0261885646 |
1.919935 |
AGGAGC,GAGGAG,GCCAAG,GGAGCC,GGAGGA |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| SRSF9 |
11 |
20254 |
0.10810811 |
0.0293536261 |
1.880864 |
AGGAGC,GAUGGA,GGAGCC,GGAGGA,GGAGGC,UGAGCA,UGGAGC,UGGAGG,UGGAUU |
AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| HNRNPA2B1 |
4 |
8607 |
0.04504505 |
0.0124747476 |
1.852358 |
AGGAGC,GCCAAG,GGAGCC |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| RBMX |
1 |
4925 |
0.01801802 |
0.0071387787 |
1.335691 |
AAGUGU |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPH3 |
2 |
7688 |
0.02702703 |
0.0111429292 |
1.278274 |
GGAGGA |
AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| SRSF2 |
33 |
88240 |
0.30630631 |
0.1278792060 |
1.260193 |
AGCAGU,AGGAGC,AGGAGG,AGUAGU,AUGGAG,CAGUAG,CGAGGA,CUCGUG,GAGCAG,GAGGAG,GAUGGA,GCAGUA,GCUACU,GGAGCC,GGAGGA,GUAGUC,GUCCCU,GUCGCC,GUGAGC,GUUCGU,UCCAGA,UCCCUG,UGGAGG,UGGCAG,UUCGUG |
AAAAGA,AAAGAG,AAAGGA,AACCAG,AACGAG,AAGAAG,AAGAGA,AAGCAG,AAGCGG,AAGCUG,AAGGAG,AAGGCG,AAGUAC,AAUACC,AAUCCC,AAUGCC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACCCGC,ACCCGG,ACCGGU,ACCGUA,ACCUCC,ACCUGC,ACGAGA,ACGAGG,ACGCUG,ACGGAG,ACGUGU,ACUACC,ACUACU,ACUAGA,ACUCAA,ACUCCC,ACUCCU,ACUGUA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGACUA,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCCC,AGCCGA,AGCCUC,AGCGGA,AGCGUG,AGCUCC,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGA,AGGCGU,AGGCUG,AGGGUA,AGUACG,AGUAGA,AGUAGG,AGUAGU,AGUAUC,AGUCGA,AGUGAC,AGUGGU,AGUGUU,AGUUCC,AUACCG,AUCACC,AUCACU,AUCCCC,AUCCCG,AUCCCU,AUCCGG,AUCGAG,AUCGCU,AUGAAU,AUGAUG,AUGCCG,AUGCGU,AUGCUG,AUGGAG,AUUACC,AUUACU,AUUAGU,AUUCCA,AUUCCC,AUUCCU,AUUGAU,CACCUC,CAGAGA,CAGAGG,CAGAGU,CAGCUA,CAGUAG,CAGUCC,CAGUGG,CAGUGU,CCACCA,CCACCG,CCACUA,CCACUG,CCAGAA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCACA,CCCCCA,CCCCCG,CCCCUA,CCCCUG,CCCGCA,CCCGCU,CCCGUG,CCCUUG,CCGAGA,CCGCAG,CCGCUA,CCGGAU,CCGGUG,CCUCAG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGAA,CGAGAG,CGAGAU,CGAGCA,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGG,CGCAGU,CGCCCU,CGCUGC,CGGAGA,CGGAGU,CGUAAG,CGUGAU,CGUGCG,CGUGUA,CGUGUU,CUAACG,CUACCA,CUACCG,CUACUA,CUACUG,CUAGAA,CUAGAC,CUCAAG,CUCCAA,CUCCCA,CUCCCG,CUCCUA,CUCCUG,CUCGAG,CUCGUG,CUGAUG,CUGCAA,CUGCUA,CUGUUA,GAAAGG,GAACCA,GAACGA,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GAAUGC,GACCAC,GACCCC,GACCCG,GACCGG,GACCGU,GACCUG,GACGCU,GACGGA,GACUAC,GACUAG,GACUCA,GACUCC,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGG,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCGU,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUAU,GAGUGA,GAUCAC,GAUCCC,GAUCCG,GAUCGA,GAUCGC,GAUGAU,GAUGCG,GAUGCU,GAUGGA,GAUUAC,GAUUAG,GAUUCC,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCAAA,GCCACC,GCCACU,GCCCAC,GCCCCC,GCCCCU,GCCCUU,GCCGAG,GCCGCA,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGAGC,GCGAGU,GCGCAG,GCGGAG,GCGGUC,GCGUGU,GCUACC,GCUACU,GCUCCA,GCUCCC,GCUCCU,GCUCGA,GCUCGU,GCUGAU,GCUGCA,GCUGCU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGACUG,GGAGAA,GGAGAG,GGAGAU,GGAGCC,GGAGCG,GGAGGA,GGAGUA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAA,GGCCAC,GGCCCA,GGCCCC,GGCCGC,GGCCUC,GGCGAG,GGCGUG,GGCUAC,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUAAG,GGUACG,GGUAGG,GGUAUG,GGUCAC,GGUCAG,GGUCCC,GGUCGC,GGUGAG,GGUGAU,GGUUAA,GGUUAC,GGUUCC,GGUUGG,GGUUGU,GUAACC,GUAAGC,GUAAGU,GUACGA,GUACGC,GUAGGC,GUAGGG,GUAGUC,GUAUCC,GUAUGC,GUCACC,GUCACU,GUCAGU,GUCCCC,GUCCCU,GUCCUC,GUCGAU,GUCGCC,GUCUAA,GUGAGC,GUGAUG,GUGCAG,GUGGUC,GUGUAA,GUUAAU,GUUACC,GUUACG,GUUACU,GUUCCA,GUUCCC,GUUCCG,GUUCCU,GUUCGA,GUUCGU,GUUCUG,GUUGGC,GUUGUU,GUUUCG,UAACCA,UAAGCU,UAAGUA,UACCAG,UACGAG,UACGCU,UACGUG,UAGACU,UAGGCU,UAUCCC,UAUGCC,UAUGCU,UCACCA,UCACCG,UCACUA,UCACUG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCCCA,UCCCCG,UCCCCU,UCCCGC,UCCCGU,UCCCUA,UCCCUG,UCCGGA,UCCUCA,UCCUCC,UCCUGC,UCCUGU,UCGAGA,UCGAGU,UCGCCG,UCGCUG,UCGUAA,UCGUGC,UCUAAC,UCUGUA,UGAAUG,UGAGCU,UGAUCG,UGAUGC,UGAUGG,UGCAGA,UGCAGU,UGCCGC,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUAC,UGUUCC,UGUUCG,UUAAUG,UUACCA,UUACCG,UUACGU,UUACUA,UUACUG,UUAGUG,UUCCAG,UUCCCA,UUCCCG,UUCCCU,UUCCGG,UUCCUA,UUCCUG,UUCGAG,UUCGUG,UUCUGU,UUGAUG,UUGGCG,UUGUUG,UUUCGA |
| SRSF10 |
4 |
13860 |
0.04504505 |
0.0200874160 |
1.165076 |
GAGGAG |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| YTHDC1 |
1 |
5576 |
0.01801802 |
0.0080822104 |
1.156619 |
UGCUAC |
GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SRSF1 |
33 |
99563 |
0.30630631 |
0.1442885423 |
1.086018 |
AGACAG,AGAUGG,AGGAGC,AGGAGG,AUGGAG,CAGACA,CCCUGG,CCGAGG,CCUGGA,CGAGGA,CUCGUG,GACAGA,GAGCAG,GAGGAG,GAGGCA,GAUGGA,GCAGCA,GCCCGA,GGAGCC,GGAGGA,GGAGGC,GGAGGU,GGUGCU,UGGAGC,UGGAGG |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AACACG,AACAGC,AACCGG,AAGAAC,AAGAAG,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGAG,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACACGA,ACACGU,ACAGAA,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCCGG,ACCCGU,ACCGGA,ACCGGU,ACGAAC,ACGAAG,ACGAAU,ACGACG,ACGCGA,ACGCGC,ACGCUC,ACGGAA,ACGGAC,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGA,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCCGU,AGCGAG,AGCGGA,AGCGGG,AGCGGU,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGCGA,AGGUAA,AGUCGG,AUACGA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,AUUACG,AUUUCA,CAACCA,CAACCC,CAACCG,CAACGA,CAACGC,CAAGCA,CAAGGA,CAAUGG,CACACG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGA,CACCGG,CACGCA,CACGCU,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGCGG,CAGUCG,CAUGGU,CCAACC,CCAACG,CCAAGC,CCAAGG,CCAAUG,CCACAG,CCACCA,CCACCC,CCACCG,CCACGA,CCACGC,CCACGG,CCACUG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCAUGA,CCAUGG,CCCACC,CCCACG,CCCACU,CCCAGC,CCCAGG,CCCAUG,CCCCCA,CCCCCC,CCCCCG,CCCCGA,CCCCGC,CCCCGG,CCCGAC,CCCGCA,CCCGCC,CCCGCG,CCCGGA,CCCGGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCCUGA,CCCUGC,CCCUGG,CCGACA,CCGACC,CCGACG,CCGACU,CCGAGC,CCGAGG,CCGAUG,CCGCCA,CCGCCC,CCGCCG,CCGCGA,CCGCGC,CCGCGG,CCGCUA,CCGGAC,CCGGAG,CCGGCA,CCGGCC,CCGGCG,CCGGGA,CCGGGC,CCGGGG,CCGUCC,CCGUCG,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCA,CCUCCG,CCUCGA,CCUCGG,CCUGAC,CCUGCA,CCUGCG,CCUGGA,CCUGGG,CGAACC,CGAACG,CGAAGC,CGAAGG,CGACAG,CGACCA,CGACCC,CGACCG,CGACGA,CGACGC,CGACGG,CGAGCA,CGAGCG,CGAGGA,CGAGGC,CGAGGG,CGAUGA,CGAUGG,CGCACA,CGCACC,CGCACG,CGCAGC,CGCAGG,CGCCCA,CGCCCC,CGCCCG,CGCCGA,CGCCGC,CGCCGG,CGCCGU,CGCGAG,CGCGCA,CGCGCC,CGCGCG,CGCGCU,CGCGGA,CGCGGC,CGCGGG,CGCGGU,CGCGUC,CGCUAU,CGCUCA,CGCUCC,CGCUCG,CGCUGC,CGCUGG,CGGAAC,CGGAAG,CGGAAU,CGGACA,CGGACC,CGGACG,CGGAGC,CGGAGG,CGGCAC,CGGCAG,CGGCCA,CGGCCC,CGGCCG,CGGCGA,CGGCGC,CGGCGG,CGGGCA,CGGGCC,CGGGCG,CGGGGA,CGGGGC,CGGGGG,CGGUCC,CGGUCG,CGGUGC,CGGUGG,CGUCCA,CGUCCG,CGUCGA,CGUCGG,CGUGCA,CGUGCG,CGUGGA,CGUGGG,CUACGA,CUAGGG,CUCAGG,CUCCGG,CUCGGA,CUCGUG,CUGAAC,CUGACU,CUGAGC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAACCA,GAACGA,GAAGAA,GAAGAC,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACCCG,GACCGA,GACGAA,GACGAC,GACGAG,GACGCA,GACGCG,GACGGA,GACGGC,GACGGG,GACUGA,GACUGG,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGCGA,GAGCGG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUACG,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GAUGGU,GCACCA,GCACCG,GCACGA,GCACGG,GCACGU,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCAUAC,GCCCAC,GCCCCA,GCCCCG,GCCCGA,GCCCGG,GCCCGU,GCCGCA,GCCGCG,GCCGGA,GCCGGG,GCCGGU,GCCGUA,GCGAGC,GCGCAA,GCGCCA,GCGCCC,GCGCCG,GCGCGA,GCGCGC,GCGCGG,GCGGAA,GCGGAC,GCGGAG,GCGGCA,GCGGCG,GCGGGA,GCGGGG,GCGGUU,GCGUCA,GCUCCA,GCUCCG,GCUCGA,GCUCGG,GCUGCA,GCUGCG,GCUGGA,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGACGG,GGACGU,GGACUG,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCC,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAC,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCACG,GGCAGA,GGCCCA,GGCCCG,GGCCGA,GGCCGG,GGCCGU,GGCGCA,GGCGCG,GGCGGA,GGCGGG,GGCGGU,GGGACG,GGGCCA,GGGCCG,GGGCGA,GGGCGG,GGGGAA,GGGGAC,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGGGG,GGGUAC,GGUAAC,GGUCCA,GGUCCG,GGUCGA,GGUCGG,GGUGAA,GGUGAC,GGUGAU,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,GUGACG,UAAUUU,UACGAA,UACGGA,UAGACA,UAGGAC,UAUUAC,UCAAGA,UCAGGU,UCCGGA,UCGCAC,UCGGGC,UCGUGU,UGAACA,UGAACC,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUA,UGACUG,UGAGUU,UGAUGA,UGAUGC,UGAUGG,UGGAAA,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGA,UGGUGC,UGGUGG,UGUAGG,UUAAUU,UUACGG,UUCAAG,UUGACA,UUUCAA |