circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000103227:-:16:954356:954666
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000103227:-:16:954356:954666 | ENSG00000103227 | ENST00000545827 | - | 16 | 954357 | 954666 | 310 | UCGUGGCAUUCCUGGUGGCUUUCCAUCAGAACAAGCAGCUCAUCGGUGACAGGGGGCUGCUUCCCUGCAGAGUGUUCCUGAAGAACUUCCAGCAGUACUUCCAGGACAGGACGAGCUGGGAAGUCUUCAGCUACAUGCCCACCAUCCUCUGGCUGAUGGACUGGUCAGACAUGAACUCCAACCUGGACUUGCUGGCUCUUCUCGGACUGGGCAUCUCGUCUUUCGUACUGAUCACGGGCUGCGCCAACAUGCUUCUCAUGGCUGCCCUGUGGGGCCUCUACAUGUCCCUGGUUAAUGUGGGCCAUGUCUGUCGUGGCAUUCCUGGUGGCUUUCCAUCAGAACAAGCAGCUCAUCGGUGAC | circ |
| ENSG00000103227:-:16:954356:954666 | ENSG00000103227 | ENST00000545827 | - | 16 | 954357 | 954666 | 22 | GGCCAUGUCUGUCGUGGCAUUC | bsj |
| ENSG00000103227:-:16:954356:954666 | ENSG00000103227 | ENST00000545827 | - | 16 | 954657 | 954866 | 210 | CUGCAGGUUGGCUUUUCCAGUGUUUCCCCCAAAAUCUUGAGAAAGUACUCAGACUCCUGAGUCUAGGGUUUGGCCCCAAACCGUCAGCCAGACUCUGCCUCCCCUUCCCGCCUGCCUCGCCCCGCAUUCUGAGCUGCGCCCAUGGUGUCCACAUGUGGUUUGUUCCUAGUCAUGCUUGUUUUGUCUUCUUUUUCUUCCAGUCGUGGCAUU | ie_up |
| ENSG00000103227:-:16:954356:954666 | ENSG00000103227 | ENST00000545827 | - | 16 | 954157 | 954366 | 210 | GCCAUGUCUGGUGAGUAGCAGGAAUGGGGCGGUCGGAGCUUAGGGCUUGCUGUCACUCAGCACAGGCACUGCAGGUGUCCUGGCACUUGGAAAGACUGGACGAGCGUUUAUCUUAAAUUCCUUCAGGAGGGUAUUGUCAUUUUACUUAGGCCAUGGCAGAGCGGCCAGUGCACCAGCCACAGCAGCCACCCAUCUUCAGGAUUAAAACCC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
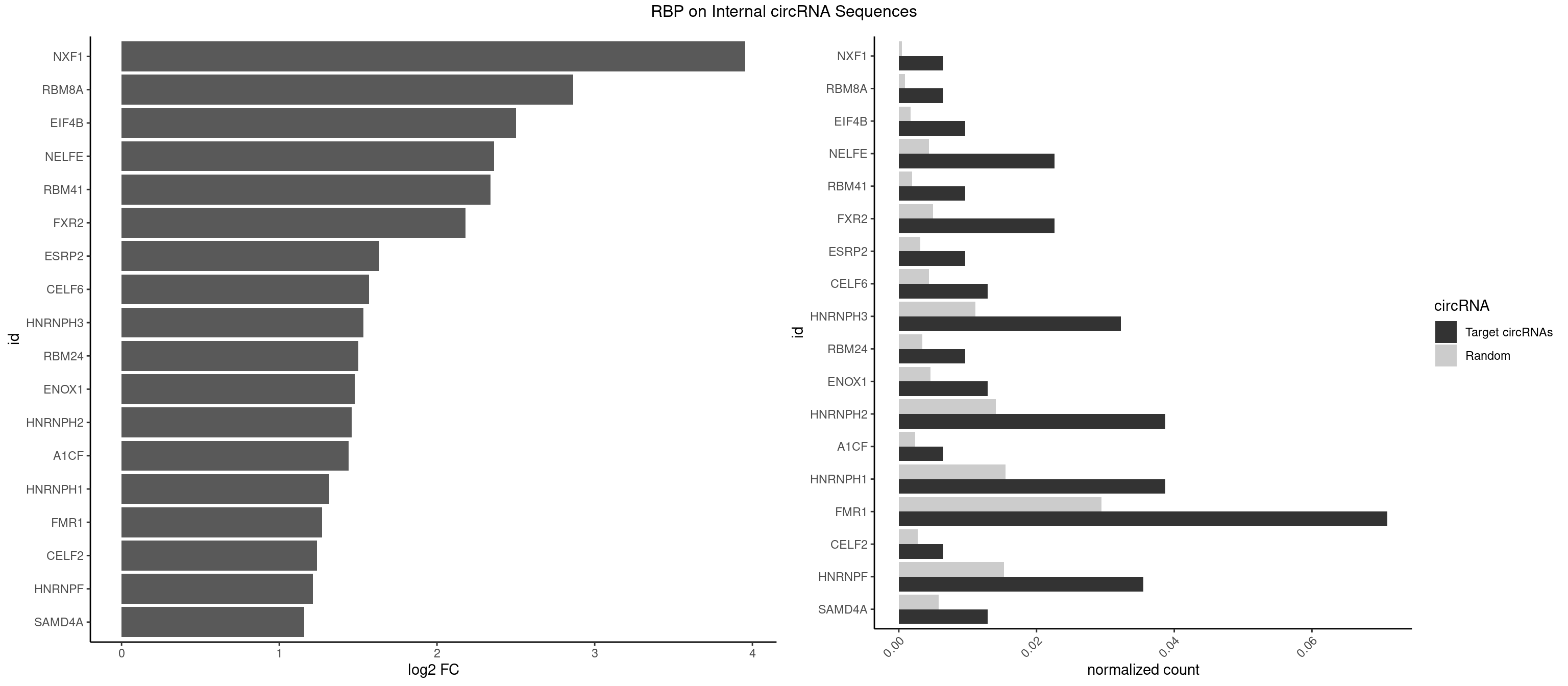
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 286 | 0.006451613 | 0.0004159215 | 3.955277 | AACCUG | AACCUG |
| RBM8A | 1 | 611 | 0.006451613 | 0.0008869128 | 2.862796 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| EIF4B | 2 | 1179 | 0.009677419 | 0.0017100607 | 2.500575 | CUCGGA,UCGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| NELFE | 6 | 3028 | 0.022580645 | 0.0043896388 | 2.362913 | CUCUGG,CUGGCU,CUGGUU,UCUGGC,UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM41 | 2 | 1318 | 0.009677419 | 0.0019115000 | 2.339917 | UACAUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| FXR2 | 6 | 3434 | 0.022580645 | 0.0049780156 | 2.181444 | GACAGG,GACGAG,GGACAG,GGACGA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ESRP2 | 2 | 2150 | 0.009677419 | 0.0031172377 | 1.634354 | GGGAAG,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF6 | 3 | 2997 | 0.012903226 | 0.0043447134 | 1.570399 | GUGGGG,UGUGGG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPH3 | 9 | 7688 | 0.032258065 | 0.0111429292 | 1.533531 | AAUGUG,AUGUGG,GGGAAG,GGGCUG,GGGGCU,GGGGGC,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| RBM24 | 2 | 2357 | 0.009677419 | 0.0034172229 | 1.501798 | AGAGUG,GAGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ENOX1 | 3 | 3195 | 0.012903226 | 0.0046316558 | 1.478132 | AGGACA,CAGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPH2 | 11 | 9711 | 0.038709677 | 0.0140746688 | 1.459593 | AAUGUG,AUGUGG,CAGGAC,GGGAAG,GGGCUG,GGGGCU,GGGGGC,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| A1CF | 1 | 1642 | 0.006451613 | 0.0023810421 | 1.438067 | UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPH1 | 11 | 10717 | 0.038709677 | 0.0155325680 | 1.317398 | AAUGUG,AUGUGG,CAGGAC,GGGAAG,GGGCUG,GGGGCU,GGGGGC,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| FMR1 | 21 | 20291 | 0.070967742 | 0.0294072466 | 1.270992 | ACUGGG,ACUGGU,AGCAGC,AGCUGG,CUGGGC,CUGGUG,GACAGG,GACUGG,GCAGCU,GCUGGG,GGACAG,GGCUGA,GGGCAU,GGGCUG,GUGGCU,UGACAG,UGGCUG | AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| CELF2 | 1 | 1886 | 0.006451613 | 0.0027346479 | 1.238305 | GUCUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| HNRNPF | 10 | 10561 | 0.035483871 | 0.0153064921 | 1.213020 | AAUGUG,AUGUGG,GGGAAG,GGGCUG,GGGGCU,GGGGGC,UGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| SAMD4A | 3 | 3992 | 0.012903226 | 0.0057866714 | 1.156926 | CUGGAC,CUGGUC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
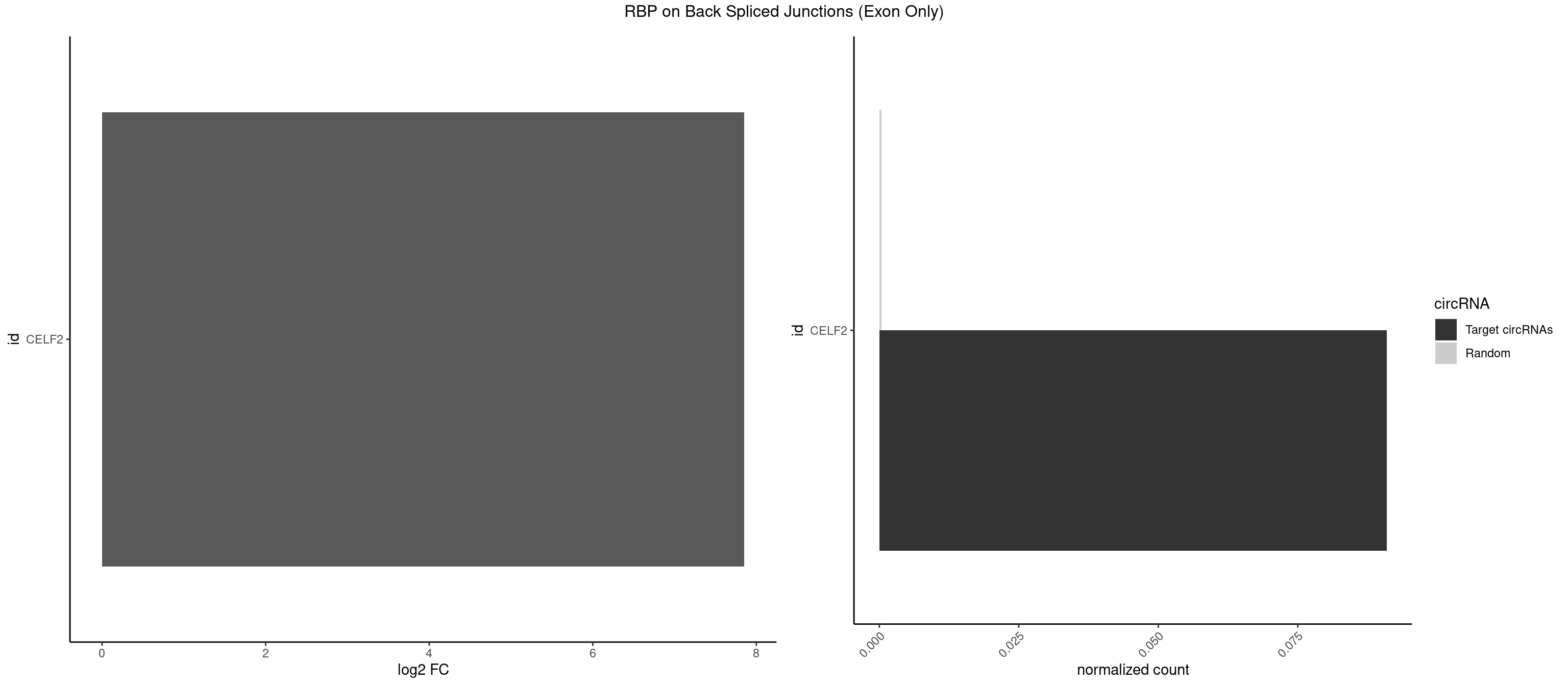
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF2 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GUCUGU | AUGUGU,GUCUGU,UAUGUU,UGUGUG,UGUUGU |
RBP on BSJ (Exon and Intron)
Plot
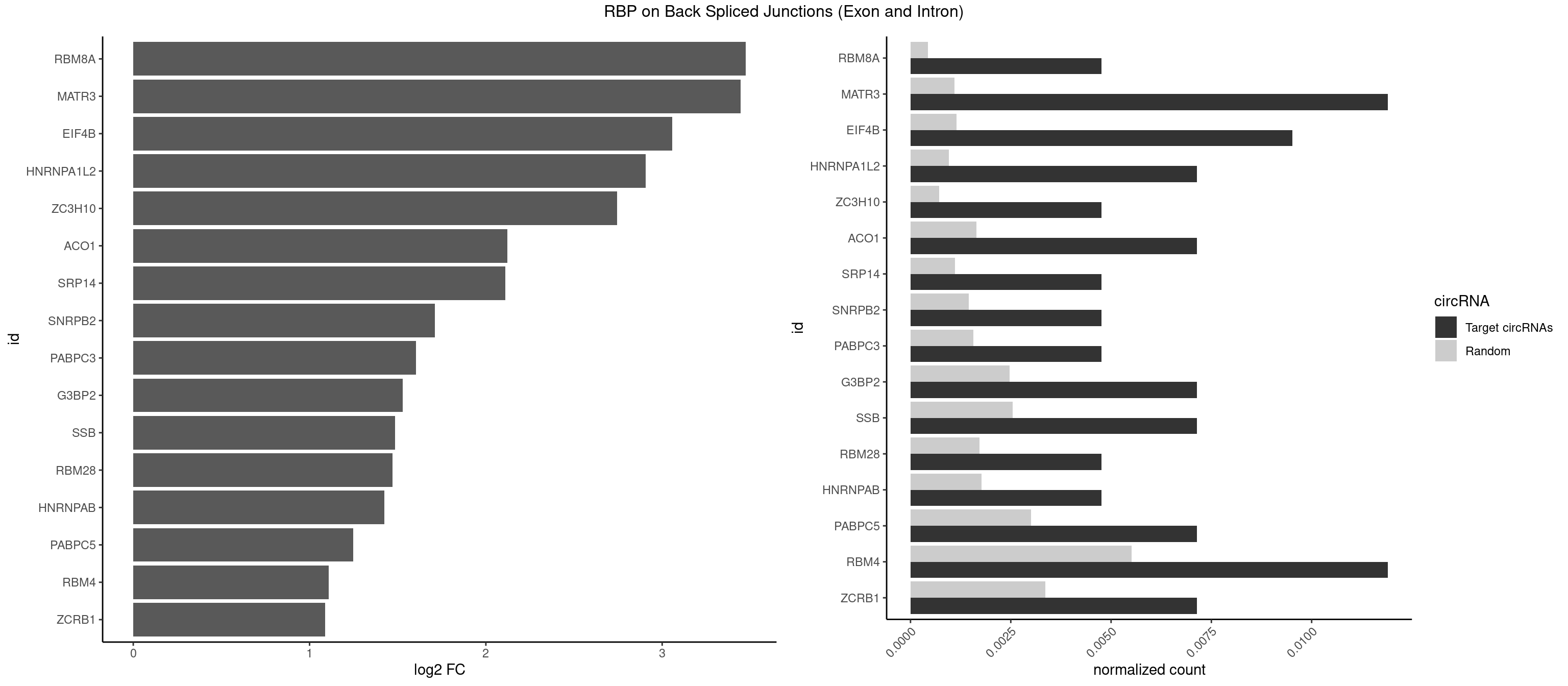
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| MATR3 | 4 | 216 | 0.011904762 | 0.0010933091 | 3.444765 | AAUCUU,AUCUUA,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| EIF4B | 3 | 226 | 0.009523810 | 0.0011436921 | 3.057840 | CUUGGA,GUCGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGU,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | CGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUU,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUUCC,CUUCUU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.