circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000258539:-:10:124681606:124706887
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000258539:-:10:124681606:124706887 | ENSG00000258539 | ENST00000494792 | - | 10 | 124681607 | 124706887 | 1080 | GGAGUUGACCACAUUUGGCCAUUUCCCAGAAGGGCCCCACCCCAAGGGUGAGUGGCCAAUGGGGAGCUGUUUCUGCUGACAUCAAUUCCCCAGGAGGUACUCACCCCAAGUCUGCCCAAGUGAAGAUGGCUGAUACCCACCCUGGGAUGGAGCCCAGCGCCUGAGGCCCUUAUCAUGGUGAUGGUCCUAAGUGAAAGCCUCAGCACCCGGGGAGCUGACUCCAUUGCAUGUGGGACCUUCAGCCGUGAACUGCACACGCCAAAGAAGAUGAGUCAAGGACCUACACUUUUCUCUUGUGGAAUUAUGGAAAAUGACAGAUGGCGAGACCUGGACAGGAAAUGCCCUCUUCAGAUUGACCAACCGAGCACCAGCAUCUGGGAAUGCCUGCCUGAAAAGGACAGCUCACUAUGGCACCGGGAGGCAGUGACCGCCUGCGCUGUGACCAGUCUGAUCAAAGACCUCAGCAUCAGCGACCACAACGGGAACCCCUCAGCACCCCCUAGCAAGCGCCAGUGCCGCUCACUGUCCUUCUCCGAUGAGAUGUCCAGUUGCCGGACAUCAUGGAGGCCCUUGGGCUCCAAAGUCUGGACUCCCGUGGAAAAGAGACGCUGCUACAGCGGGGGCAGCGUCCAGCGCUAUUCCAACGGCUUCAGCACCAUGCAGAGGAGUUCCAGCUUCAGCCUCCCUUCCCGGGCCAACGUGCUCUCCUCACCCUGCGACCAGGCAGGACUCCACCACCGAUUUGGAGGGCAGCCCUGCCAAGGGGUGCCAGGCUCAGCCCCGUGUGGACAGGCAGGUGACACCUGGAGCCCUGACCUGCACCCCGUGGGAGGAGGCCGGCUGGACCUGCAGCGGUCCCUCUCUUGCUCACAUGAGCAGUUUUCCUUUGUGGAAUACUGUCCUCCCUCAGCCAACAGCACACCUGCCUCAACACCAGAGCUGGCGAGACGCUCCAGCGGCCUUUCCCGCAGCCGCUCCCAGCCGUGUGUCCUUAACGACAAGAAGGUCGGUGUUAAAAGGCGGCGCCCUGAAGAAGUGCAAGAGCAGAGGCCUUCUCUAGACCUUGCCAAGAUGGCACAGGGAGUUGACCACAUUUGGCCAUUUCCCAGAAGGGCCCCACCCCAAGGGUG | circ |
| ENSG00000258539:-:10:124681606:124706887 | ENSG00000258539 | ENST00000494792 | - | 10 | 124681607 | 124706887 | 22 | AGAUGGCACAGGGAGUUGACCA | bsj |
| ENSG00000258539:-:10:124681606:124706887 | ENSG00000258539 | ENST00000494792 | - | 10 | 124706878 | 124707087 | 210 | CCCAAGGCUGGCAUGGCAGGGGUGGGCGGGGGGACUCUGAGAAGGUAACUCUGGGCCUUGGUGACUCUCCCAGAGUGCAUCUCCUUUAGCAGAGAGGUGGUUCCAGUCCCCUGGGAUUUCCAGUACUGUGGGAGGGGUGACUCUCAGGGCCCUCGUCUUUCUUAGUCCUGAAUCUUUUUGCUUUUCUUUUGUCUUUCCAGGGAGUUGACC | ie_up |
| ENSG00000258539:-:10:124681606:124706887 | ENSG00000258539 | ENST00000494792 | - | 10 | 124681407 | 124681616 | 210 | GAUGGCACAGGUAAGAGCCCCAGCAGCCUGGAGUCACCUUGGUGCUCACUGGAGCCUCUGCUGGUUCUGGGCCGUCCUAGACAAGCAUAGAUUGCCAGUUUCAGAAAUACCCAGGAGUGGGUUAAUUUCCUUGUUUUUUUAAAUCCUAUUCAAAGAAAGAUUAUUUCCAGUUGGUUUUUAAAAAGCGUUUAUUUCUAUUUACAAGCAAAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
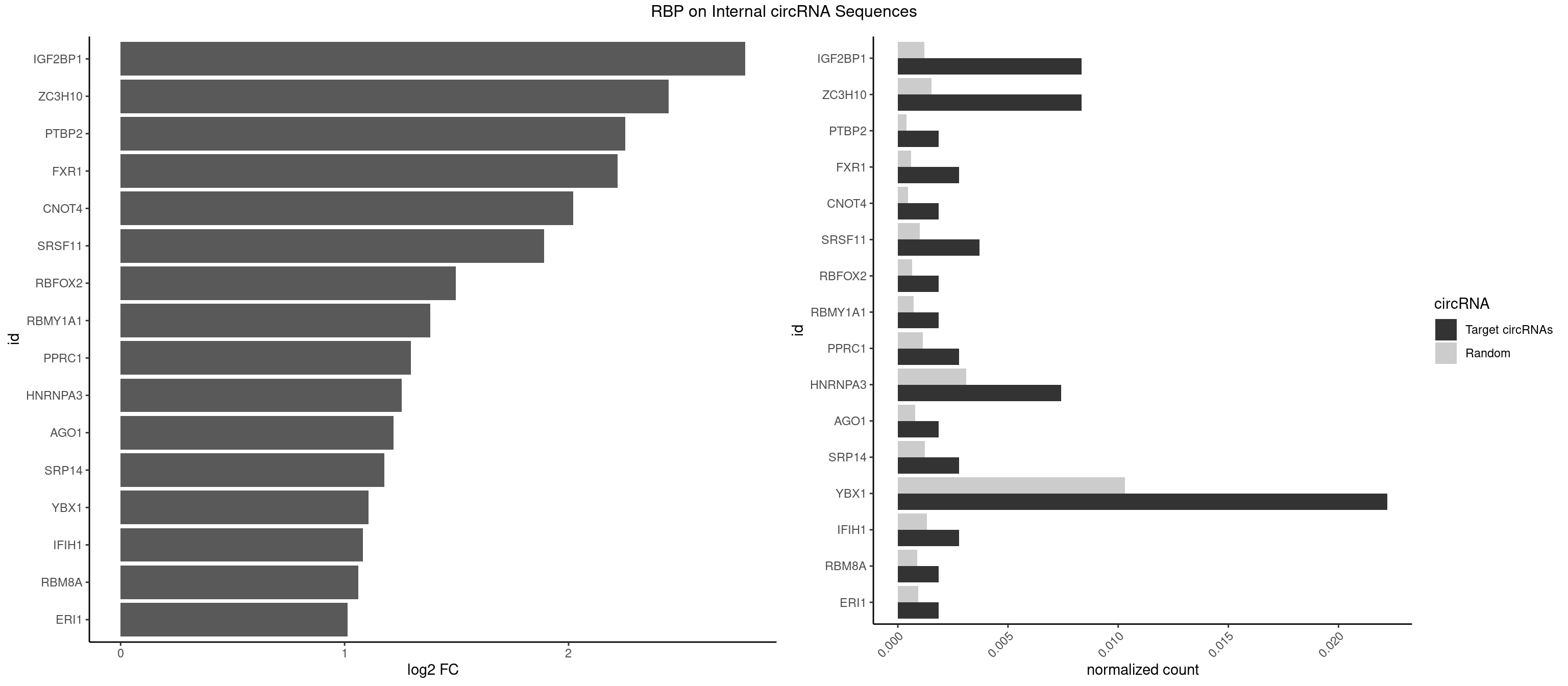
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGF2BP1 | 8 | 831 | 0.008333333 | 0.0012057377 | 2.788978 | AGCACC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ZC3H10 | 8 | 1053 | 0.008333333 | 0.0015274610 | 2.447758 | CAGCGA,CAGCGC,CCAGCG,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PTBP2 | 1 | 267 | 0.001851852 | 0.0003883867 | 2.253403 | CUCUCU | CUCUCU |
| FXR1 | 2 | 411 | 0.002777778 | 0.0005970720 | 2.217954 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| CNOT4 | 1 | 314 | 0.001851852 | 0.0004564992 | 2.020284 | GACAGA | GACAGA |
| SRSF11 | 3 | 688 | 0.003703704 | 0.0009985015 | 1.891132 | AAGAAG | AAGAAG |
| RBFOX2 | 1 | 452 | 0.001851852 | 0.0006564894 | 1.496125 | UGCAUG | UGACUG,UGCAUG |
| RBMY1A1 | 1 | 489 | 0.001851852 | 0.0007101099 | 1.382854 | ACAAGA | ACAAGA,CAAGAC |
| PPRC1 | 2 | 780 | 0.002777778 | 0.0011318283 | 1.295276 | CGGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| HNRNPA3 | 7 | 2140 | 0.007407407 | 0.0031027457 | 1.255423 | CAAGGA,CCAAGG,GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| AGO1 | 1 | 548 | 0.001851852 | 0.0007956130 | 1.218830 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 847 | 0.002777778 | 0.0012289250 | 1.176534 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| YBX1 | 23 | 7119 | 0.022222222 | 0.0103183321 | 1.106793 | ACACCA,ACAUCA,ACCACA,ACCACC,CACACC,CACCAC,CAUCAU,CAUCUG,CCACAA,CCACCA,CCAGCA,CCCUGC,CCUGCG,GCCUGC,GUCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| IFIH1 | 2 | 904 | 0.002777778 | 0.0013115296 | 1.082681 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 1 | 611 | 0.001851852 | 0.0008869128 | 1.062104 | UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| ERI1 | 1 | 632 | 0.001851852 | 0.0009173461 | 1.013431 | UUCAGA | UUCAGA,UUUCAG |
RBP on BSJ (Exon Only)
Plot
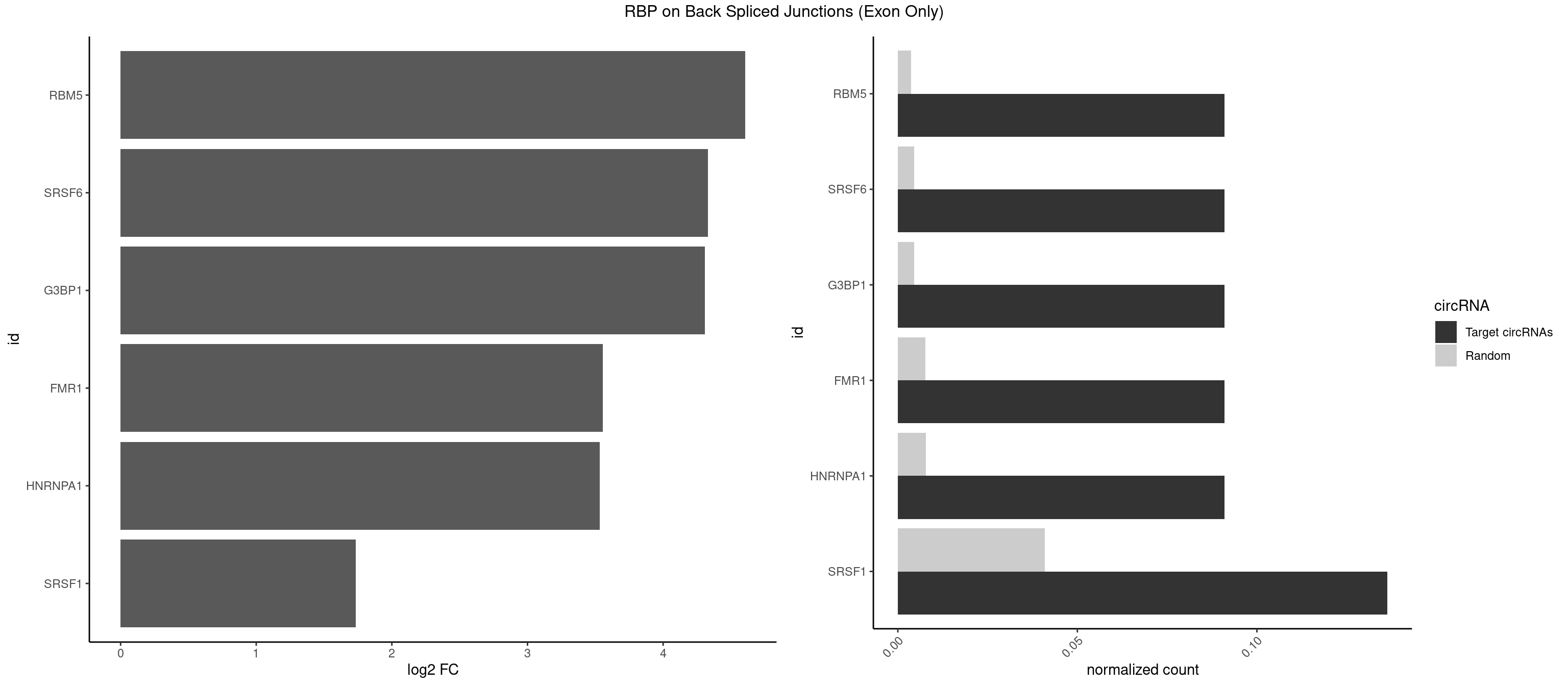
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | AGGGAG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | CACAGG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
| G3BP1 | 1 | 69 | 0.09090909 | 0.004584752 | 4.309509 | CACAGG | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | AGGGAG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | CAGGGA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| SRSF1 | 2 | 625 | 0.13636364 | 0.041000786 | 1.733736 | ACAGGG,CACAGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
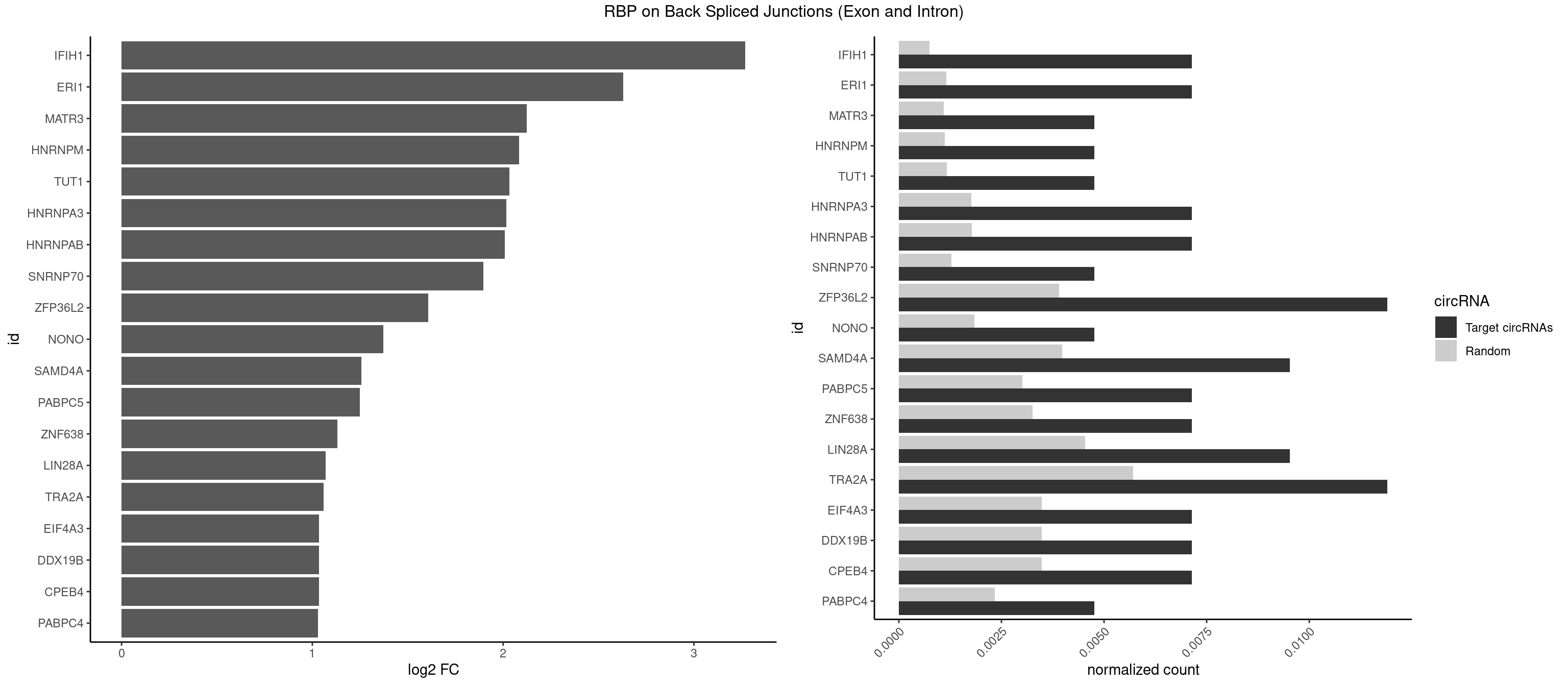
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IFIH1 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | GGCCCU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CCAAGG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AGACAA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGCA,GCUGGC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | AGGAGU,GGAGGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAAGAA,AAGAAA,AGAAAG,GAAAGA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.