circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000144741:+:3:66262050:66346408
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000144741:+:3:66262050:66346408 | ENSG00000144741 | MSTRG.23213.14 | + | 3 | 66262051 | 66346408 | 198 | GUUGCCUGCCUGAUUCGAGUUCCAUCUGAAGUGGUUAAGCAGAGGGCACAGGUAUCUGCUUCUACAAGAACAUUUCAGAUUUUCUCUAACAUCUUAUAUGAAGAGGGUAUCCAAGGGUUGUAUCGAGGCUAUAAAAGCACAGUUUUAAGAGAGAUUCCUUUUUCUUUGGUCCAGUUUCCCUUAUGGGAGUCCUUAAAAGUUGCCUGCCUGAUUCGAGUUCCAUCUGAAGUGGUUAAGCAGAGGGCACA | circ |
| ENSG00000144741:+:3:66262050:66346408 | ENSG00000144741 | MSTRG.23213.14 | + | 3 | 66262051 | 66346408 | 22 | AGUCCUUAAAAGUUGCCUGCCU | bsj |
| ENSG00000144741:+:3:66262050:66346408 | ENSG00000144741 | MSTRG.23213.14 | + | 3 | 66261851 | 66262060 | 210 | GAAAGGUUCUGGGCUGACAAAACAAUGUACAUGCUUUAAGGGUCCAAGAGUUUUAGGUUAAAAAAAAAAAAAGAAACUUUUUGACAUCUUGCAGUAAGCCUACAUAUUAUACCUUUUUACUACAAUUUUUGCUUACCAAAAAAUUUCAAUUUUUAUAGCAGUUAUUUACUUUUUAGGAUAUAAUCAAAUUUGUCUUUUAGGUUGCCUGCC | ie_up |
| ENSG00000144741:+:3:66262050:66346408 | ENSG00000144741 | MSTRG.23213.14 | + | 3 | 66346399 | 66346608 | 210 | GUCCUUAAAAGUAAGUUUCUCAGUCUUCACAUAAAAUUUGUCUCUAAUUAUAACAUACCUAAUAGUUUGAGUGUGGAAGAGUAGAACAUAAUGUUGACUAGAAGUUCAGCAACUCAAACCUAUUAGCAGUUGAAAAUAUAUUUUAUGUGUGAUUUACAUGCCAUUUCAGCCUUUUAAACUUGUUCAGGUGGCUGUGCUCCUAGGAAAUAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
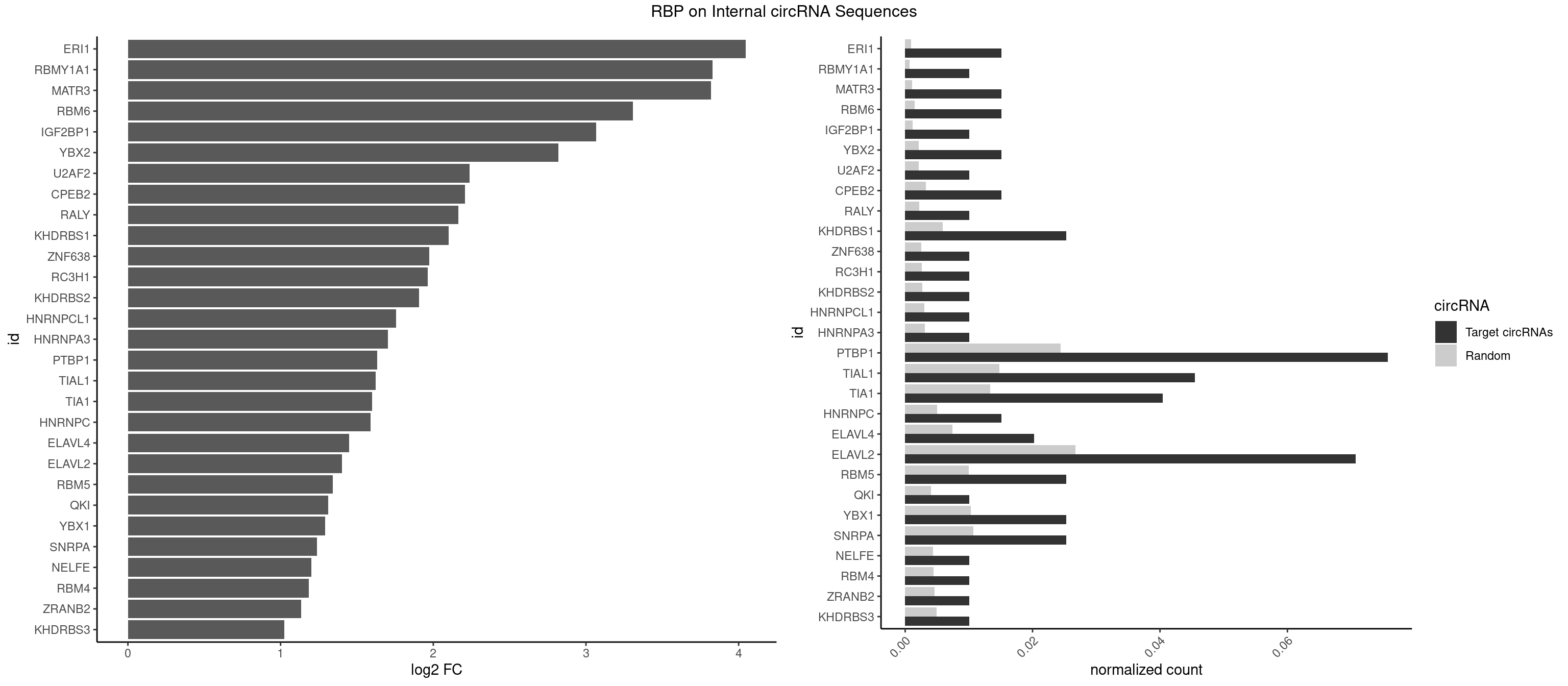
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ERI1 | 2 | 632 | 0.01515152 | 0.0009173461 | 4.045852 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBMY1A1 | 1 | 489 | 0.01010101 | 0.0007101099 | 3.830313 | ACAAGA | ACAAGA,CAAGAC |
| MATR3 | 2 | 739 | 0.01515152 | 0.0010724109 | 3.820532 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM6 | 2 | 1054 | 0.01515152 | 0.0015289102 | 3.308887 | AUCCAA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| IGF2BP1 | 1 | 831 | 0.01010101 | 0.0012057377 | 3.066512 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 1480 | 0.01515152 | 0.0021462711 | 2.819558 | AACAUC,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| U2AF2 | 1 | 1477 | 0.01010101 | 0.0021419234 | 2.237521 | UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| CPEB2 | 2 | 2261 | 0.01515152 | 0.0032780993 | 2.208531 | CCUUUU,CUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY | 1 | 1553 | 0.01010101 | 0.0022520629 | 2.165181 | UUUUUC | UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 4 | 4064 | 0.02525253 | 0.0058910141 | 2.099840 | AUAAAA,UAAAAG,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ZNF638 | 1 | 1773 | 0.01010101 | 0.0025708878 | 1.974161 | GGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 1 | 1786 | 0.01010101 | 0.0025897275 | 1.963627 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| KHDRBS2 | 1 | 1858 | 0.01010101 | 0.0026940701 | 1.906640 | AUAAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPCL1 | 1 | 2062 | 0.01010101 | 0.0029897078 | 1.756423 | CUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPA3 | 1 | 2140 | 0.01010101 | 0.0031027457 | 1.702882 | CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PTBP1 | 14 | 16854 | 0.07575758 | 0.0244263326 | 1.632953 | AUUUUC,CAUCUU,CCAUCU,CCUUUU,CUUUUU,UCCAUC,UUCCUU,UUCUCU,UUCUUU,UUUCCC,UUUCUU,UUUUCU,UUUUUC | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| TIAL1 | 8 | 10182 | 0.04545455 | 0.0147572438 | 1.623001 | AGUUUU,AUUUUC,CAGUUU,CUUUUU,UUUCAG,UUUUAA,UUUUUC | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 | 7 | 9206 | 0.04040404 | 0.0133428208 | 1.598436 | AUUUUC,CUUUUU,GUUUUA,UCCUUU,UUUUCU,UUUUUC | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPC | 2 | 3471 | 0.01515152 | 0.0050316361 | 1.590363 | CUUUUU,UUUUUC | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 3 | 5105 | 0.02020202 | 0.0073996354 | 1.448973 | GUAUCU,UGUAUC,UUGUAU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL2 | 13 | 18468 | 0.07070707 | 0.0267653478 | 1.401488 | AUUUUC,CUUAUA,CUUUUU,GAUUUU,GUUUUA,UCUUAU,UUAUAU,UUCUUU,UUGUAU,UUUAAG,UUUCUU,UUUUAA,UUUUUC | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM5 | 4 | 6879 | 0.02525253 | 0.0099705232 | 1.340687 | AGGGUA,CAAGGG,GAGGGU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| QKI | 1 | 2805 | 0.01010101 | 0.0040664663 | 1.312652 | CUAACA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| YBX1 | 4 | 7119 | 0.02525253 | 0.0103183321 | 1.291218 | AACAUC,ACAUCU,CAUCUG,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| SNRPA | 4 | 7380 | 0.02525253 | 0.0106965744 | 1.239279 | AUUCCU,GAUUCC,GGGUAU,GUUUCC | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| NELFE | 1 | 3028 | 0.01010101 | 0.0043896388 | 1.202325 | UGGUUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 1 | 3065 | 0.01010101 | 0.0044432593 | 1.184809 | UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZRANB2 | 1 | 3173 | 0.01010101 | 0.0045997733 | 1.134865 | AGGUAU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| KHDRBS3 | 1 | 3429 | 0.01010101 | 0.0049707696 | 1.022958 | AUAAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
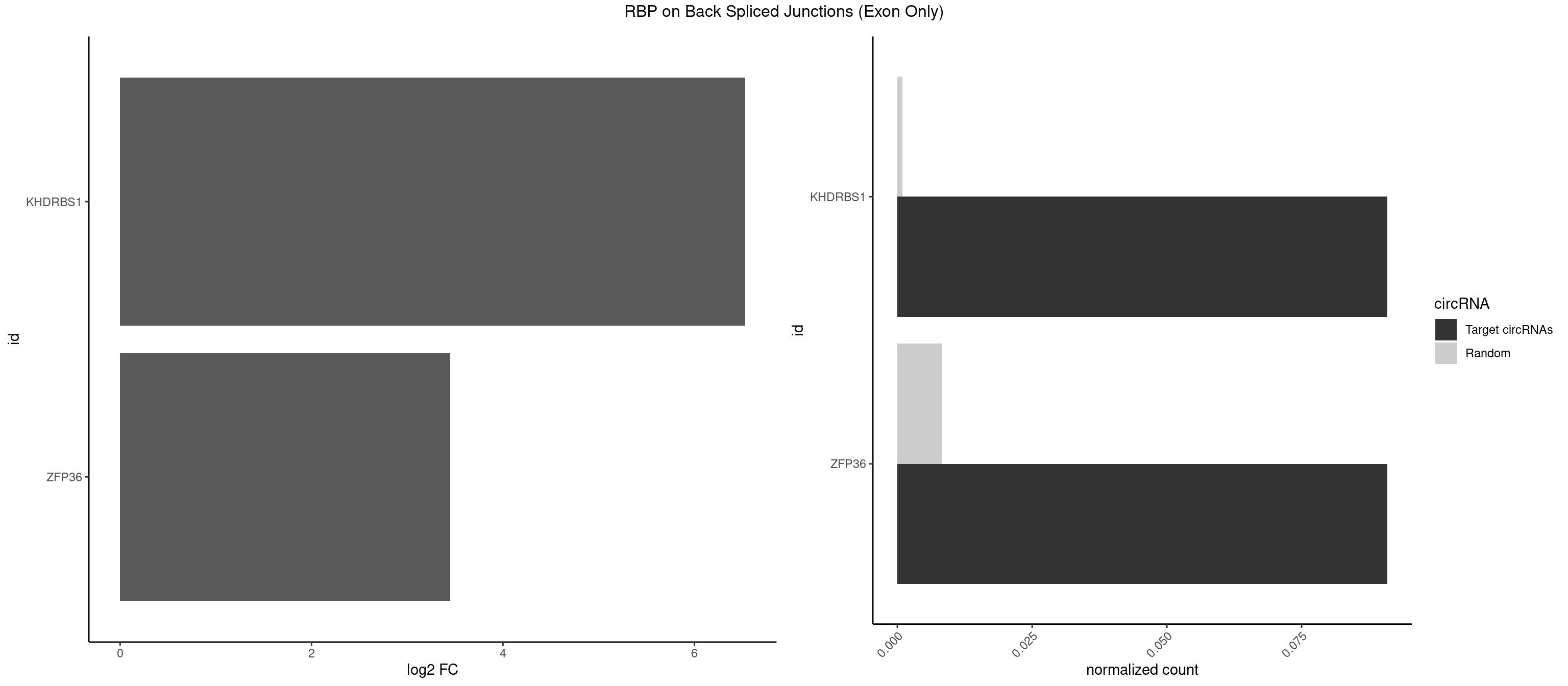
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| KHDRBS1 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | UAAAAG | AUUUAA,CUAAAA,GAAAAC,UAAAAA,UAAAAG,UUAAAA,UUAAAU |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | AAAAGU | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
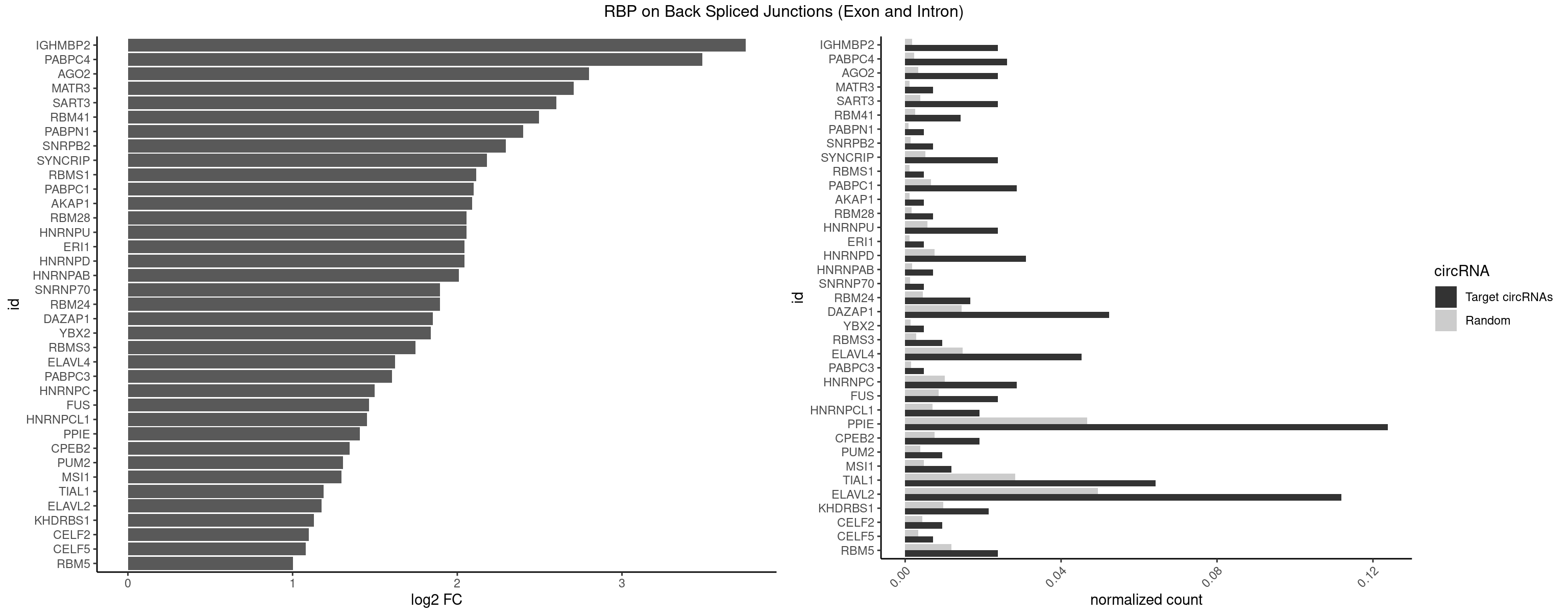
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGHMBP2 | 9 | 350 | 0.023809524 | 0.0017684401 | 3.750989 | AAAAAA | AAAAAA |
| PABPC4 | 10 | 462 | 0.026190476 | 0.0023327287 | 3.488952 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| AGO2 | 9 | 677 | 0.023809524 | 0.0034159613 | 2.801175 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SART3 | 9 | 777 | 0.023809524 | 0.0039197904 | 2.602690 | AAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | UACAUG,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| SYNCRIP | 9 | 1041 | 0.023809524 | 0.0052498992 | 2.181177 | AAAAAA | AAAAAA,UUUUUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| PABPC1 | 11 | 1321 | 0.028571429 | 0.0066606207 | 2.100845 | AAAAAA,CAAACC,CUAAUA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGA,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPU | 9 | 1136 | 0.023809524 | 0.0057285369 | 2.055300 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPD | 12 | 1488 | 0.030952381 | 0.0075020153 | 2.044700 | AAAAAA,UAUUUA,UUAGGA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AUAGCA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGUGUG,GAGUGU,GUGUGA,GUGUGG,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| DAZAP1 | 21 | 2878 | 0.052380952 | 0.0145052398 | 1.852468 | AAAAAA,AGGAAA,AGGAUA,AGGUUA,AGGUUG,AGUAAG,AGUUUG,UAGGAA,UAGGAU,UAGGUU,UAGUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AAUAUA,AUAUAU,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ELAVL4 | 18 | 2916 | 0.045238095 | 0.0146966949 | 1.622046 | AAAAAA,UAUUUA,UAUUUU,UCUAAU,UUAUUU,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPC | 11 | 2006 | 0.028571429 | 0.0101118501 | 1.498526 | AUUUUU,CUUUUU,GGAUAU,UUUUUA,UUUUUG | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FUS | 9 | 1711 | 0.023809524 | 0.0086255542 | 1.464850 | AAAAAA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPCL1 | 7 | 1381 | 0.019047619 | 0.0069629182 | 1.451847 | AUUUUU,CUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PPIE | 51 | 9262 | 0.123809524 | 0.0466696896 | 1.407565 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAU,AAAUUU,AAUAUA,AAUUAU,AAUUUU,AUAAAA,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAUA,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAUUA,UAUAAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAUAA,UUAUUU,UUUAAA,UUUAUA,UUUUAA,UUUUAU,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | AUUUUU,CCUUUU,CUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUACAU,UACAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUGG,AGUAAG,UAGGAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| TIAL1 | 26 | 5597 | 0.064285714 | 0.0282043531 | 1.188580 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUU,CUUUUA,CUUUUU,UAUUUU,UUAUUU,UUUAAA,UUUCAG,UUUUAA,UUUUAU,UUUUUA,UUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 46 | 9826 | 0.111904762 | 0.0495112858 | 1.176442 | AAUUUG,AAUUUU,AUAUUU,AUUUAC,AUUUUA,AUUUUU,CUUUAA,CUUUUA,CUUUUU,GAUUUA,GUUUUA,UAAGUU,UACUUU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCUUUU,UGAUUU,UUACUU,UUAUAC,UUAUGU,UUAUUU,UUUAAG,UUUACU,UUUAUA,UUUAUG,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 | 8 | 1945 | 0.021428571 | 0.0098045143 | 1.128018 | AUAAAA,AUUUAC,UAAAAA,UAAAAG,UUAAAA,UUUUAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AAAAAA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.