circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000117114:+:1:81836884:81907230
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000117114:+:1:81836884:81907230 | ENSG00000117114 | ENST00000674492 | + | 1 | 81836885 | 81907230 | 387 | AUAUGAAGAUCAAUGAUGCAGACUGAUGGUUUUGAUGAAGCUGGGCAUUUAUAACUAGAUUCAUUAAGGAAUACAAAGAAAAUACUUAAAGGGAUCAAUAAUGGUGUCUUCUGGUUGCAGAAUGCGAAGUCUGUGGUUUAUCAUUGUAAUCAGCUUCUUACCAAAUACAGAAGGUUUCAGCAGAGCAGCUUUACCAUUUGGGCUGGUGAGGCGAGAAUUAUCCUGUGAAGGUUAUUCUAUAGAUCUGCGAUGCCCGGGCAGUGAUGUCAUCAUGAUUGAGAGCGCUAACUAUGGUCGGACGGAUGACAAGAUUUGUGAUGCUGACCCAUUUCAGAUGGAGAAUACAGACUGCUACCUCCCCGAUGCCUUCAAAAUUAUGACUCAAAGAUAUGAAGAUCAAUGAUGCAGACUGAUGGUUUUGAUGAAGCUGGGCAUUU | circ |
| ENSG00000117114:+:1:81836884:81907230 | ENSG00000117114 | ENST00000674492 | + | 1 | 81836885 | 81907230 | 22 | AUGACUCAAAGAUAUGAAGAUC | bsj |
| ENSG00000117114:+:1:81836884:81907230 | ENSG00000117114 | ENST00000674492 | + | 1 | 81836685 | 81836894 | 210 | AAGGCAGUCAACCUUUUUGGGCUUUUGACUGUUUUCAGAAUGUAAGAUUCAGAUAUCCAGUGUAUAGGAGCAGCUUCCAUAGUGGACAUAGAAUCAAAAUAUGUUUUAUAUUCAGGAGAGAGAGAGAGGAACGGAGAGAGAGAAUUGUUUUGUUAUGUAGAAAGACCAUAGAGUAAGUGAUGGAUUUGUUUGUUUUUCAGAUAUGAAGAU | ie_up |
| ENSG00000117114:+:1:81836884:81907230 | ENSG00000117114 | ENST00000674492 | + | 1 | 81907221 | 81907430 | 210 | UGACUCAAAGGUAAAUAUUCAUGUGUUAAUGUCCCAUUUGAGCUAUUGUAUCCAAAUUAGAAAAAUUAUUACAGUUAUUCCAAAGCGAAUGGAUAUAGACCACAUCCCAGCCUAUUUAUUAAACUGAGUUGGGUUUUUACCUACUAAUGUUGAUAUCUACACUUUAUAUUUUCAUCAACAUUCAAAUAUGUAAAUCAUUCUAAUAUAGCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
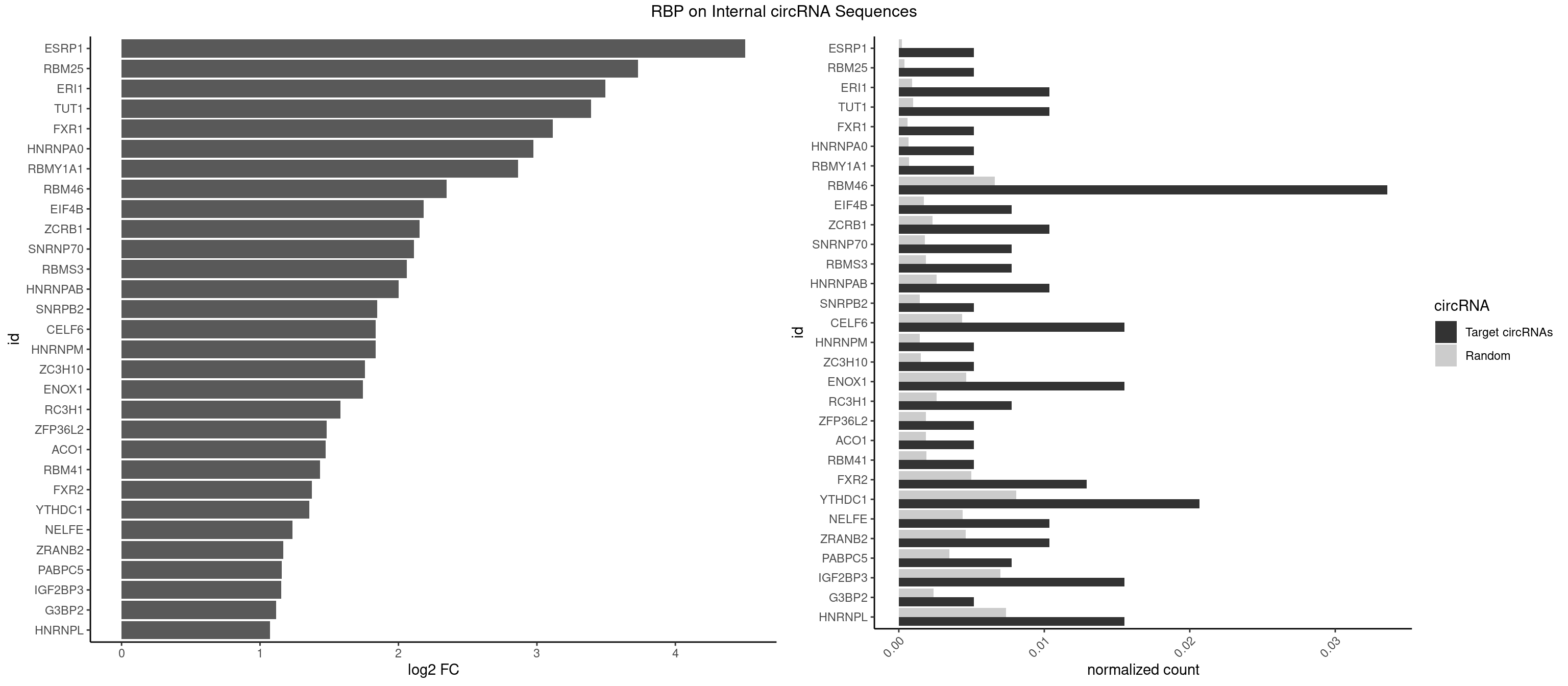
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.005167959 | 0.0002275250 | 4.505497 | AGGGAU | AGGGAU |
| RBM25 | 1 | 268 | 0.005167959 | 0.0003898359 | 3.728656 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| ERI1 | 3 | 632 | 0.010335917 | 0.0009173461 | 3.494056 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 3 | 678 | 0.010335917 | 0.0009840095 | 3.392850 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| FXR1 | 1 | 411 | 0.005167959 | 0.0005970720 | 3.113618 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPA0 | 1 | 453 | 0.005167959 | 0.0006579386 | 2.973570 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.005167959 | 0.0007101099 | 2.863480 | ACAAGA | ACAAGA,CAAGAC |
| RBM46 | 12 | 4554 | 0.033591731 | 0.0066011240 | 2.347323 | AAUGAU,AUCAAU,AUCAUG,AUCAUU,AUGAAG,AUGAUG,AUGAUU,GAUCAA,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| EIF4B | 2 | 1179 | 0.007751938 | 0.0017100607 | 2.180510 | GUCGGA,UCGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZCRB1 | 3 | 1605 | 0.010335917 | 0.0023274215 | 2.150862 | ACUUAA,GAAUUA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SNRNP70 | 2 | 1237 | 0.007751938 | 0.0017941145 | 2.111285 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMS3 | 2 | 1283 | 0.007751938 | 0.0018607779 | 2.058651 | CUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPAB | 3 | 1782 | 0.010335917 | 0.0025839306 | 2.000027 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRPB2 | 1 | 991 | 0.005167959 | 0.0014376103 | 1.845922 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF6 | 5 | 2997 | 0.015503876 | 0.0043447134 | 1.835296 | GUGAGG,GUGAUG,UGUGAU,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPM | 1 | 999 | 0.005167959 | 0.0014492040 | 1.834334 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 1 | 1053 | 0.005167959 | 0.0015274610 | 1.758459 | GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ENOX1 | 5 | 3195 | 0.015503876 | 0.0046316558 | 1.743029 | AAUACA,AUACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RC3H1 | 2 | 1786 | 0.007751938 | 0.0025897275 | 1.581757 | CUUCUG,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZFP36L2 | 1 | 1277 | 0.005167959 | 0.0018520827 | 1.480446 | AUUUAU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ACO1 | 1 | 1283 | 0.005167959 | 0.0018607779 | 1.473689 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 1 | 1318 | 0.005167959 | 0.0019115000 | 1.434889 | AUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| FXR2 | 4 | 3434 | 0.012919897 | 0.0049780156 | 1.375952 | GACAAG,GACGGA,GGACGG,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| YTHDC1 | 7 | 5576 | 0.020671835 | 0.0080822104 | 1.354845 | GAAUAC,GAAUGC,GACUGC,UGAUGC,UGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| NELFE | 3 | 3028 | 0.010335917 | 0.0043896388 | 1.235492 | CUGGUU,GCUAAC,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZRANB2 | 3 | 3173 | 0.010335917 | 0.0045997733 | 1.168032 | AGGUUA,AGGUUU,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| PABPC5 | 2 | 2400 | 0.007751938 | 0.0034795387 | 1.155661 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 5 | 4815 | 0.015503876 | 0.0069793662 | 1.151461 | AAAUAC,AAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| G3BP2 | 1 | 1644 | 0.005167959 | 0.0023839405 | 1.116246 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPL | 5 | 5085 | 0.015503876 | 0.0073706513 | 1.072765 | AAAUAC,AAUACA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
RBP on BSJ (Exon Only)
Plot
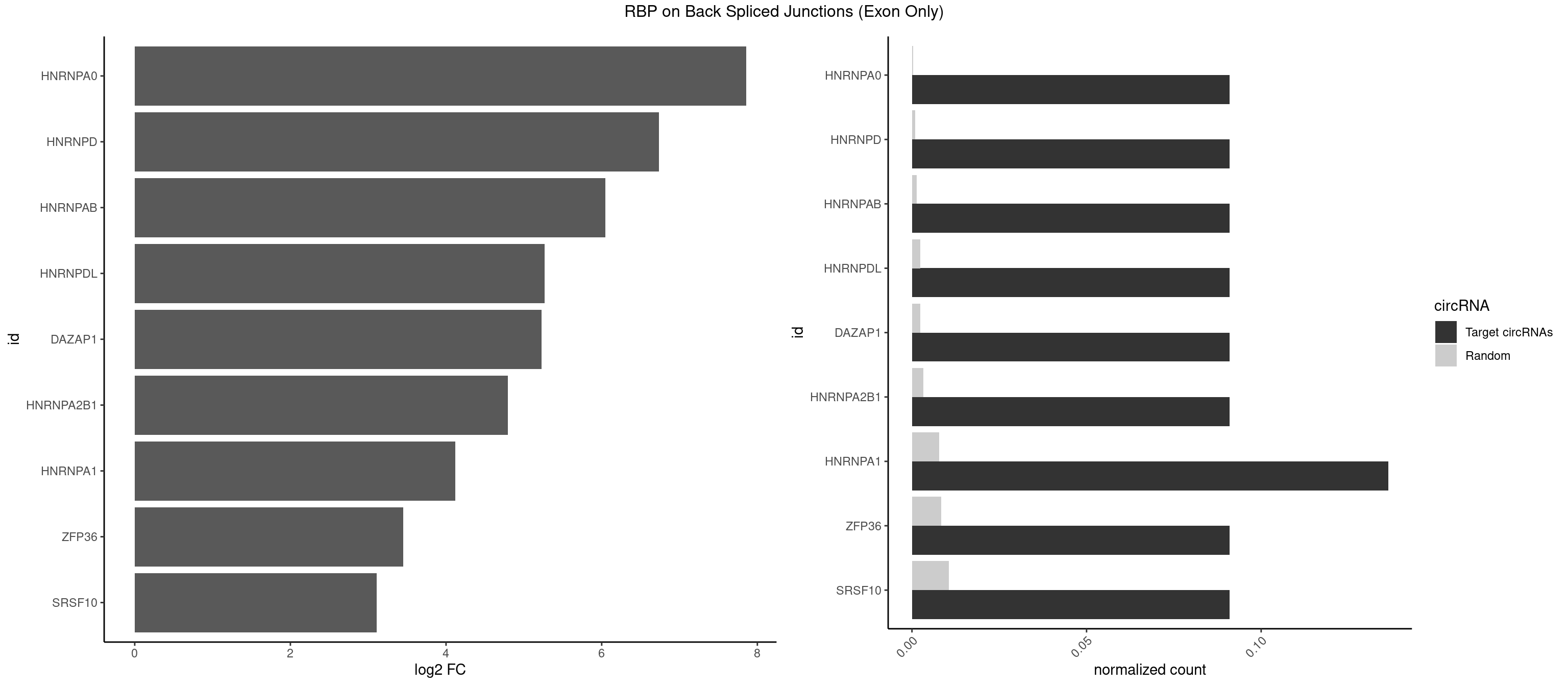
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| HNRNPD | 1 | 12 | 0.09090909 | 0.0008514540 | 6.738352 | AGAUAU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,UUAGAG,UUAGGA,UUAGGG |
| HNRNPAB | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA |
| HNRNPDL | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | AGAUAU | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGAUAU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | AGAUAU | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| HNRNPA1 | 2 | 119 | 0.13636364 | 0.0078595756 | 4.116864 | AGAUAU,CAAAGA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | CAAAGA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | CAAAGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
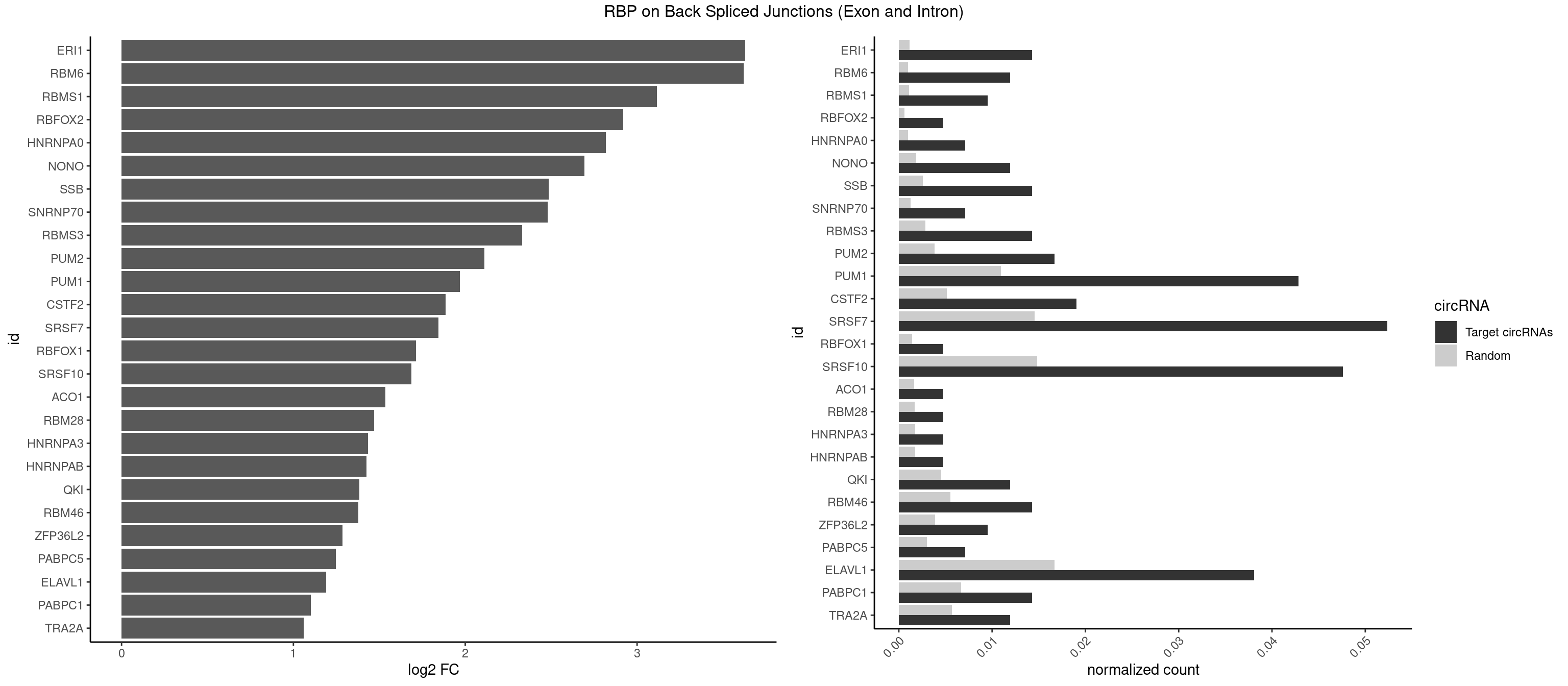
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ERI1 | 5 | 228 | 0.014285714 | 0.0011537686 | 3.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBM6 | 4 | 191 | 0.011904762 | 0.0009673519 | 3.621354 | AUCCAA,AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| NONO | 4 | 364 | 0.011904762 | 0.0018389762 | 2.694564 | AGAGGA,AGGAAC,GAGAGG,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SSB | 5 | 505 | 0.014285714 | 0.0025493753 | 2.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AAUCAA,AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMS3 | 5 | 562 | 0.014285714 | 0.0028365578 | 2.332360 | AAUAUA,AUAUAG,UAUAGA,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PUM2 | 6 | 764 | 0.016666667 | 0.0038542926 | 2.112428 | GUAAAU,UAAAUA,UGUAAA,UGUAGA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PUM1 | 17 | 2172 | 0.042857143 | 0.0109482064 | 1.968841 | AAUAUU,AAUGUU,AAUUGU,AGAAUU,AUUGUA,CAGAAU,GAAUUG,GUAAAU,UAAAUA,UAAUAU,UAAUGU,UGUAAA,UGUAGA,UGUAUA,UUAAUG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUUUUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SRSF7 | 21 | 2893 | 0.052380952 | 0.0145808142 | 1.844971 | AGAGAA,AGAGAG,AGAGGA,AUAGAA,AUAGAC,CGAAUG,GAGAGA,GAGGAA,UCAACA,UGGACA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRSF10 | 19 | 2937 | 0.047619048 | 0.0148024990 | 1.685698 | AAAGAC,AGAGAA,AGAGAG,AGAGGA,GAAAGA,GAGAGA,GAGAGG,GAGGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AGGAGC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | AAUCAU,ACUAAU,AUCUAC,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AAUCAA,AAUCAU,AUCAAA,AUCAUU,AUGAAG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | AUUUAU,UAUUUA,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,AGAAAG | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | AUUUAU,UAUUUA,UAUUUU,UGUUUU,UUGUUU,UUUAUU,UUUGUU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACUAAU,AGAAAA,CAAAUA,CUAAUA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AGAAAG,AGAGGA,GAAAGA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.