circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000155313:+:21:15808808:15849872
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000155313:+:21:15808808:15849872 | ENSG00000155313 | ENST00000400183 | + | 21 | 15808809 | 15849872 | 1767 | CAAGAUGUGAGUGAGUUUACACACAAAUUAUUAGAUUGGUUAGAAGAUGCCUUCCAAAUGAAAGCUGAAGAGGAGACGGAUGAAGAGAAGCCAAAGAACCCCAUGGUAGAGUUGUUCUAUGGCAGAUUCCUGGCUGUGGGAGUACUUGAAGGUAAAAAAUUUGAAAACACUGAAAUGUUUGGUCAGUACCCACUUCAGGUCAAUGGGUUCAAAGAUCUGCAUGAGUGCCUAGAAGCUGCAAUGAUUGAAGGAGAAAUUGAGUCUUUACAUUCAGAGAAUUCAGGAAAAUCAGGCCAAGAGCAUUGGUUUACUGAAUUACCACCUGUGUUAACAUUUGAAUUGUCAAGAUUUGAAUUUAAUCAGGCAUUGGGAAGACCAGAAAAAAUUCACAACAAAUUAGAAUUUCCCCAAGUUUUAUAUUUGGACAGAUACAUGCACAGAAACAGAGAAAUAACAAGAAUUAAGAGGGAAGAGAUCAAGAGACUGAAAGAUUACCUCACGGUAUUACAACAAAGGCUAGAAAGAUAUUUAAGCUAUGGUUCCGGUCCCAAACGAUUCCCCUUGGUAGAUGUUCUUCAGUAUGCAUUGGAAUUUGCCUCAAGUAAACCUGUUUGCACUUCUCCUGUUGACGAUAUUGACGCUAGUUCCCCACCUAGUGGUUCCAUACCAUCACAGACAUUACCAAGCACAACAGAACAACAGGGAGCCCUAUCUUCAGAACUGCCAAGCACAUCACCUUCAUCAGUUGCUGCCAUUUCAUCGAGAUCAGUAAUACACAAACCAUUUACUCAGUCCCGGAUACCUCCAGAUUUGCCCAUGCAUCCGGCACCAAGGCACAUAACGGAGGAAGAACUUUCUGUGCUGGAAAGUUGUUUACAUCGCUGGAGGACAGAAAUAGAAAAUGACACCAGAGAUUUGCAGGAAAGCAUAUCCAGAAUCCAUCGAACAAUUGAAUUAAUGUACUCUGACAAAUCUAUGAUACAAGUUCCUUAUCGAUUACAUGCCGUUUUAGUUCACGAAGGCCAAGCUAAUGCUGGGCACUACUGGGCAUAUAUUUUUGAUCAUCGUGAAAGCAGAUGGAUGAAGUACAAUGAUAUUGCUGUGACAAAAUCAUCAUGGGAAGAGCUAGUGAGGGACUCUUUUGGUGGUUAUAGAAAUGCCAGUGCAUACUGUUUAAUGUACAUAAAUGAUAAGGCACAGUUCCUAAUACAAGAGGAGUUUAAUAAAGAAACUGGGCAGCCCCUUGUUGGUAUAGAAACAUUACCACCGGAUUUGAGAGAUUUUGUUGAGGAAGACAACCAACGAUUUGAAAAAGAACUAGAAGAAUGGGAUGCACAACUUGCCCAGAAAGCUUUGCAGGAAAAGCUUUUAGCGUCUCAGAAAUUGAGAGAGUCAGAGACUUCUGUGACAACAGCACAAGCAGCAGGAGACCCAGAAUAUCUAGAGCAGCCAUCAAGAAGUGAUUUCUCAAAGCACUUGAAAGAAGAAACUAUUCAAAUAAUUACCAAGGCAUCACAUGAGCAUGAAGAUAAAAGUCCUGAAACAGUUUUGCAGUCGAAACCUGAAAAUACUACAAGCCAACCACUUUCUAAUCAGCGAGUUGUAGAGGUGGCGAUCCCUCAUGUAGGGAAAUUUAUGAUUGAAUCAAAGGAGGGGGGGUAUGAUGACGAGAUCAUGAUGACACCGAACAUGCAAGGUAUUAUCAUGGCGAUAGGUAAAUCCAGGAGUGUAUAUGACAGGUGUGGCCCUGAAGCAGGGUUCUUUAAGCAAGAUGUGAGUGAGUUUACACACAAAUUAUUAGAUUGGUUAGAAGAUGC | circ |
| ENSG00000155313:+:21:15808808:15849872 | ENSG00000155313 | ENST00000400183 | + | 21 | 15808809 | 15849872 | 22 | GGUUCUUUAAGCAAGAUGUGAG | bsj |
| ENSG00000155313:+:21:15808808:15849872 | ENSG00000155313 | ENST00000400183 | + | 21 | 15808609 | 15808818 | 210 | AAUGCUUGUUUUUUCAAAUAUCUUAUAUCCUGGUUAAAUUAUAAAAGUUUCUUGCAUUUCGUGUGAAACAUUUCACUUGUUAUCUUUGUAAGGAGUUGGUAAUUAUAUAUUUCAACAACUGGCUCUAGGUUUGAUGUUACAACAUCUAGUAUUUUUUUUUUAGUAGGCUCAAAUAUCAACAGGCUUUUUUCUUUUCACAGCAAGAUGUGA | ie_up |
| ENSG00000155313:+:21:15808808:15849872 | ENSG00000155313 | ENST00000400183 | + | 21 | 15849863 | 15850072 | 210 | GUUCUUUAAGGUACAAUGAACAUUUUCAUUUUCGUGUCUUUUCUUUUUCAAAAUUAAACUUCUUAAAAUAUUUUAUUUCUUUGAAUUCAGUUUGGUUUUCUGUAUAAUUUCUCUUUUCCUAUCUUAGUUAUGGCCACCGGAUAUCCUAGGGUUUUAUUUUAUGUAUUAAUUAACCUAAUACUGAUUGUUUUUCUCUUAAUCCAGGUAAUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
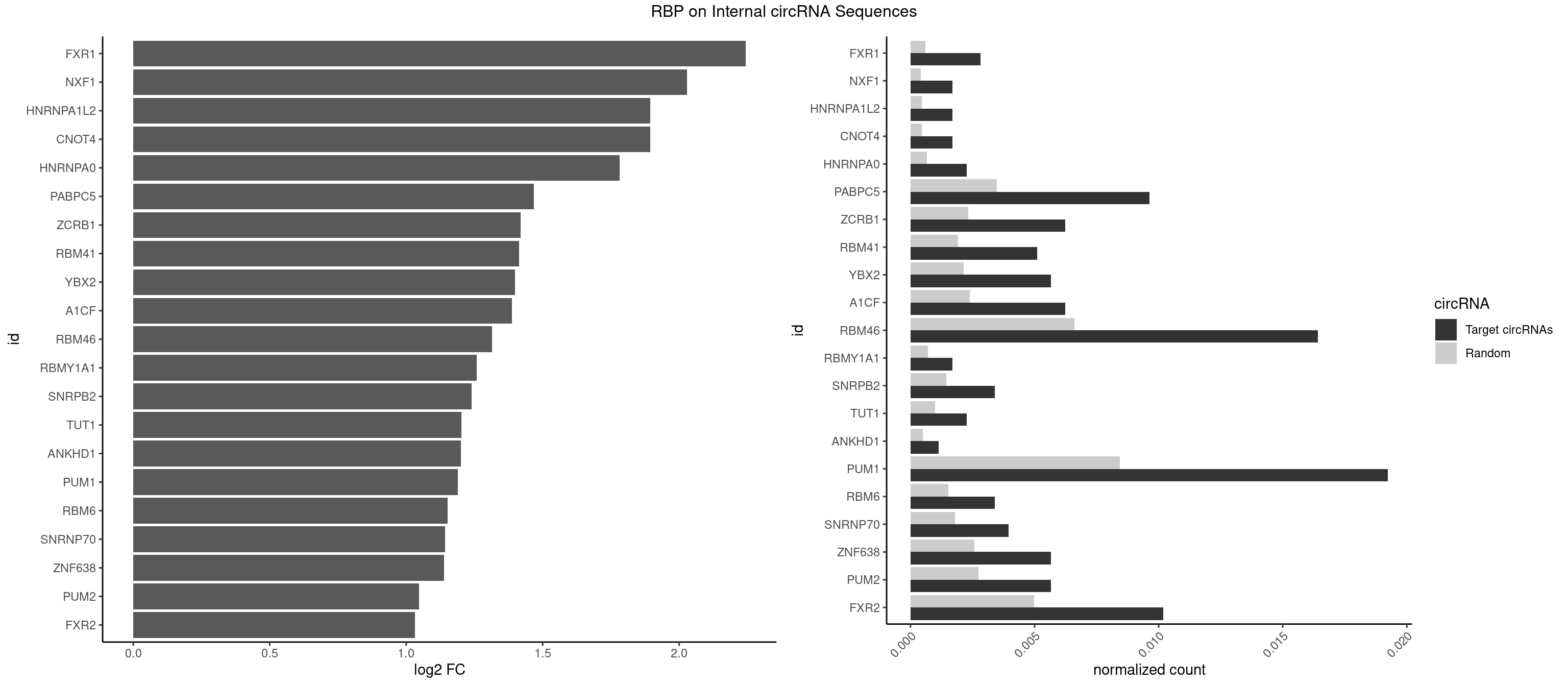
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 4 | 411 | 0.002829655 | 0.0005970720 | 2.244649 | AUGACA,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| NXF1 | 2 | 286 | 0.001697793 | 0.0004159215 | 2.029277 | AACCUG | AACCUG |
| CNOT4 | 2 | 314 | 0.001697793 | 0.0004564992 | 1.894976 | GACAGA | GACAGA |
| HNRNPA1L2 | 2 | 314 | 0.001697793 | 0.0004564992 | 1.894976 | GUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 3 | 453 | 0.002263724 | 0.0006579386 | 1.782673 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| PABPC5 | 16 | 2400 | 0.009620826 | 0.0034795387 | 1.467265 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZCRB1 | 10 | 1605 | 0.006225241 | 0.0023274215 | 1.419397 | AAUUAA,AUUUAA,GAAUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM41 | 8 | 1318 | 0.005093379 | 0.0019115000 | 1.413918 | AUACAU,UACAUG,UACAUU,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX2 | 9 | 1480 | 0.005659310 | 0.0021462711 | 1.398794 | AACAAC,ACAACA,ACAACU,ACAUCA,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| A1CF | 10 | 1642 | 0.006225241 | 0.0023810421 | 1.386536 | AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UCAGUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM46 | 28 | 4554 | 0.016411998 | 0.0066011240 | 1.313967 | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAUG,AUGAAA,AUGAAG,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMY1A1 | 2 | 489 | 0.001697793 | 0.0007101099 | 1.257546 | ACAAGA | ACAAGA,CAAGAC |
| SNRPB2 | 5 | 991 | 0.003395586 | 0.0014376103 | 1.239988 | UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TUT1 | 3 | 678 | 0.002263724 | 0.0009840095 | 1.201954 | AAAUAC,AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ANKHD1 | 1 | 339 | 0.001131862 | 0.0004927293 | 1.199831 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| PUM1 | 33 | 5823 | 0.019241653 | 0.0084401638 | 1.188890 | AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,CAGAAU,CCAGAA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,UAAUGU,UACAUA,UACAUC,UGUACA,UGUAGA,UGUAUA,UUAAUG,UUGUAG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBM6 | 5 | 1054 | 0.003395586 | 0.0015289102 | 1.151157 | AAUCCA,AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SNRNP70 | 6 | 1237 | 0.003961517 | 0.0017941145 | 1.142781 | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZNF638 | 9 | 1773 | 0.005659310 | 0.0025708878 | 1.138359 | GGUUCU,GUUCUU,GUUGGU,GUUGUU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PUM2 | 9 | 1890 | 0.005659310 | 0.0027404447 | 1.046216 | GUAAAU,GUACAU,GUAGAU,GUAUAU,UACAUA,UACAUC,UGUACA,UGUAGA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| FXR2 | 17 | 3434 | 0.010186757 | 0.0049780156 | 1.033052 | AGACAA,AGACGG,GACAAA,GACAGA,GACAGG,GACGAG,GACGGA,GGACAG,UGACAA,UGACAG,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
RBP on BSJ (Exon Only)
Plot
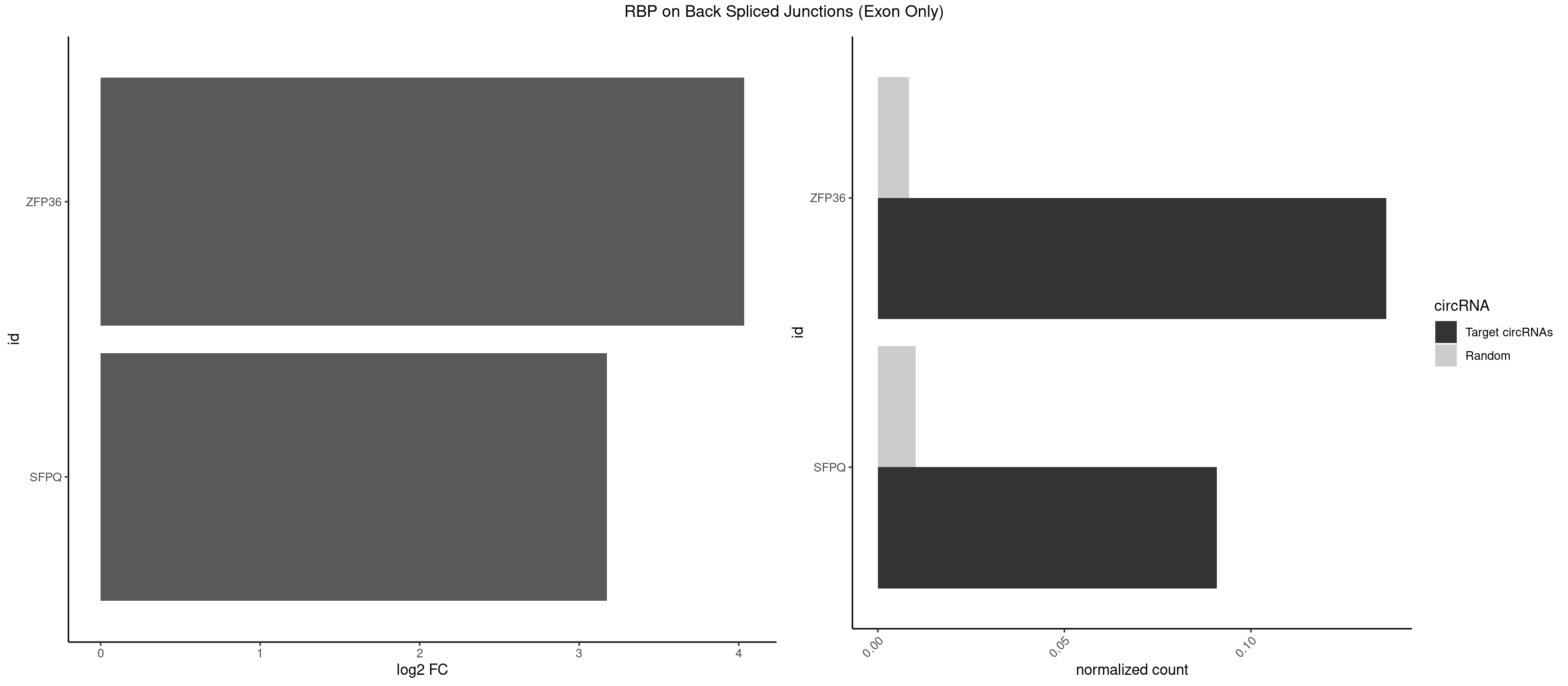
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZFP36 | 2 | 126 | 0.13636364 | 0.008318051 | 4.035070 | AAGCAA,UAAGCA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | AAGCAA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
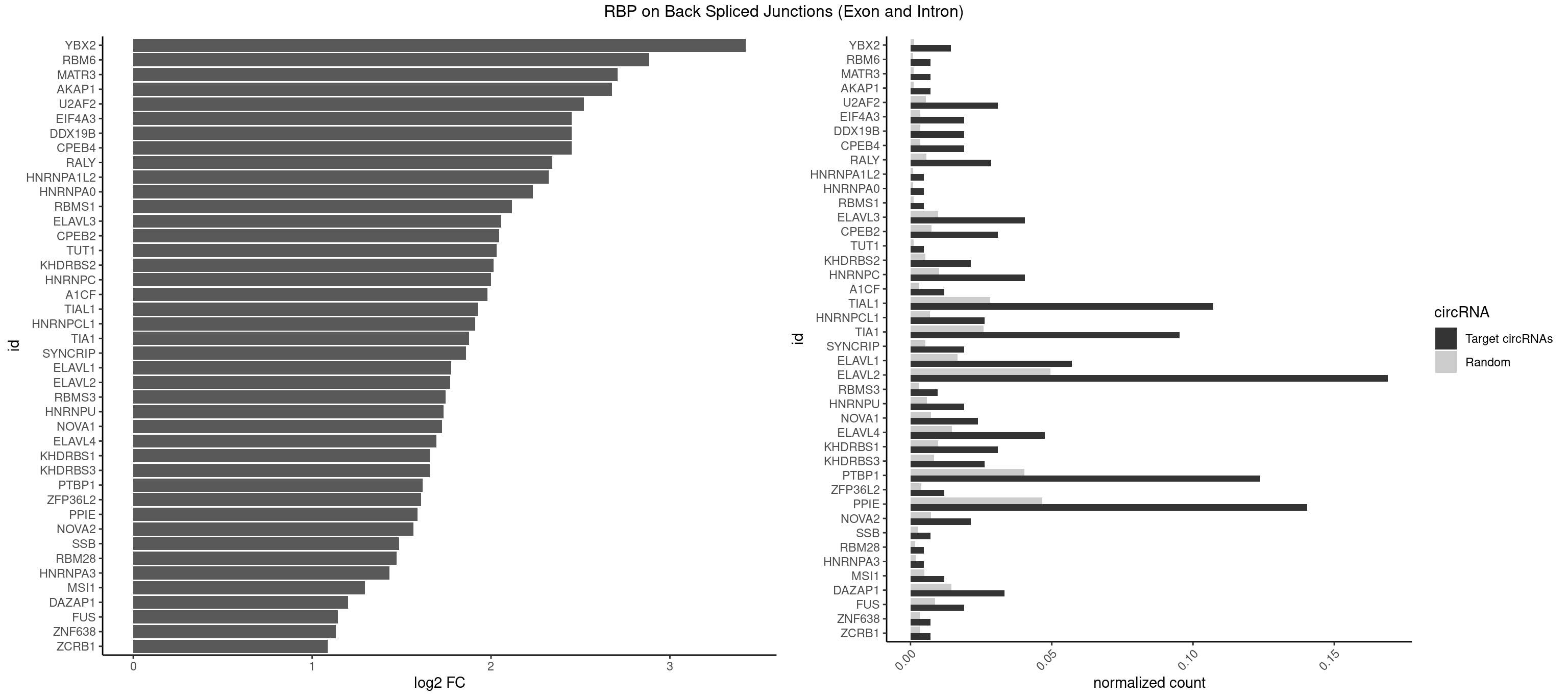
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX2 | 5 | 263 | 0.014285714 | 0.0013301088 | 3.424957 | AACAAC,AACAUC,ACAACA,ACAACU,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| U2AF2 | 12 | 1071 | 0.030952381 | 0.0054010480 | 2.518739 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| CPEB4 | 7 | 691 | 0.019047619 | 0.0034864974 | 2.449760 | UUUUUU | UUUUUU |
| DDX19B | 7 | 691 | 0.019047619 | 0.0034864974 | 2.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 7 | 691 | 0.019047619 | 0.0034864974 | 2.449760 | UUUUUU | UUUUUU |
| RALY | 11 | 1117 | 0.028571429 | 0.0056328094 | 2.342647 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ELAVL3 | 16 | 1928 | 0.040476190 | 0.0097188634 | 2.058214 | AUUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CPEB2 | 12 | 1487 | 0.030952381 | 0.0074969770 | 2.045670 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| KHDRBS2 | 8 | 1051 | 0.021428571 | 0.0053002821 | 2.015395 | AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPC | 16 | 2006 | 0.040476190 | 0.0101118501 | 2.001027 | AUUUUU,CUUUUU,GGAUAU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| TIAL1 | 44 | 5597 | 0.107142857 | 0.0282043531 | 1.925546 | AUUUUA,AUUUUC,AUUUUU,CAGUUU,CUUUUC,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAUU,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 10 | 1381 | 0.026190476 | 0.0069629182 | 1.911278 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| TIA1 | 39 | 5140 | 0.095238095 | 0.0259018541 | 1.878483 | AUUUUC,AUUUUU,CUUUUC,CUUUUU,GUUUUA,GUUUUC,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP | 7 | 1041 | 0.019047619 | 0.0052498992 | 1.859249 | UUUUUU | AAAAAA,UUUUUU |
| ELAVL1 | 23 | 3309 | 0.057142857 | 0.0166767432 | 1.776736 | UAUUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| ELAVL2 | 70 | 9826 | 0.169047619 | 0.0495112858 | 1.771600 | AUAUUU,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUUU,GUUUUA,GUUUUC,GUUUUU,UAAUUU,UAGUUA,UAUAUA,UAUAUU,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGUAUU,UUAAUU,UUAGUU,UUAUAU,UUAUGU,UUAUUU,UUCAUU,UUCUUU,UUUAAG,UUUAGU,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,UAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPU | 7 | 1136 | 0.019047619 | 0.0057285369 | 1.733372 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| NOVA1 | 9 | 1428 | 0.023809524 | 0.0071997179 | 1.725526 | AUCAAC,UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| ELAVL4 | 19 | 2916 | 0.047619048 | 0.0146966949 | 1.696047 | UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS1 | 12 | 1945 | 0.030952381 | 0.0098045143 | 1.658532 | AUAAAA,CAAAAU,UAAAAG,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| KHDRBS3 | 10 | 1646 | 0.026190476 | 0.0082980653 | 1.658195 | AUAAAA,AUUAAA,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PTBP1 | 51 | 8003 | 0.123809524 | 0.0403264813 | 1.618323 | AUUUUC,AUUUUU,CUAUCU,CUCUUA,CUCUUU,CUUCUU,CUUUUC,CUUUUU,GUCUUU,UAUUUU,UCUCUU,UCUUUU,UUAUUU,UUCUCU,UUCUUG,UUCUUU,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PPIE | 58 | 9262 | 0.140476190 | 0.0466696896 | 1.589768 | AAAAUA,AAAAUU,AAAUAU,AAAUUA,AAUAUU,AAUUAA,AAUUAU,AUAAAA,AUAAUU,AUAUAU,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUUUA,AUUUUU,UAAAAU,UAAAUU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUUU,UUAAAA,UUAAAU,UUAAUU,UUAUAA,UUAUAU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.0072299476 | 1.567479 | AUCAAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGUAGG,AGUUGG,UAGUAG,UAGUUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| DAZAP1 | 13 | 2878 | 0.033333333 | 0.0145052398 | 1.200391 | AGGUAA,AGUAGG,AGUUUG,UAGGUU,UAGUAU,UAGUUA,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| FUS | 7 | 1711 | 0.019047619 | 0.0086255542 | 1.142922 | UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.