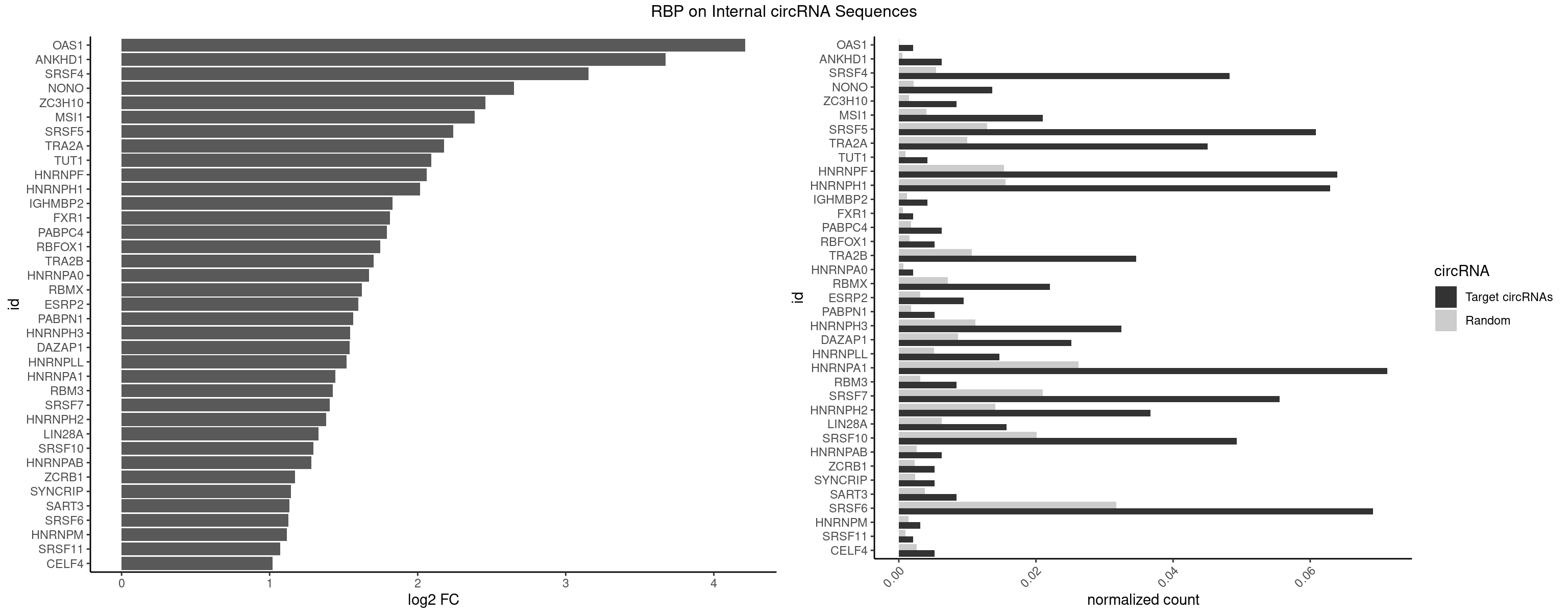
| OAS1 |
1 |
77 |
0.002096436 |
0.0001130379 |
4.213060 |
GCAUAA |
GCAUAA |
| ANKHD1 |
5 |
339 |
0.006289308 |
0.0004927293 |
3.674034 |
AGACGU,GACGUU |
AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF4 |
45 |
3740 |
0.048218029 |
0.0054214720 |
3.152816 |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| NONO |
12 |
1498 |
0.013626834 |
0.0021723567 |
2.649118 |
GAGAGG,GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ZC3H10 |
7 |
1053 |
0.008385744 |
0.0015274610 |
2.456803 |
CAGCGA,CCAGCG |
CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| MSI1 |
19 |
2770 |
0.020964361 |
0.0040157442 |
2.384200 |
AGGAAG,AGGAGG,UAGGAA |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SRSF5 |
57 |
8869 |
0.060796646 |
0.0128544391 |
2.241725 |
AACAGC,AAGAAG,AGAAGA,AGGAAG,CACAGA,CACAGC,CGCAGA,GAAGAA,GAGGAA,GGAAGA,UGCAGA,UGCAGC,UGCGGA,UGCGUC |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| TRA2A |
42 |
6871 |
0.045073375 |
0.0099589296 |
2.178213 |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGGAAG,GAAGAA,GAAGAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TUT1 |
3 |
678 |
0.004192872 |
0.0009840095 |
2.091195 |
AGAUAC,GAUACU |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPF |
60 |
10561 |
0.063941300 |
0.0153064921 |
2.062604 |
AAGGCG,AAGGGG,AGGAAG,AGGGGA,CUGGGG,GAAGGG,GAGGAA,GGAAGG,GGAGGA,GGAUGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGGAG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 |
59 |
10717 |
0.062893082 |
0.0155325680 |
2.017605 |
AAGGCG,AAGGGG,AGGAAG,AGGGGA,CUGGGG,GAAGGG,GAGGAA,GGAAGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGGAG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| IGHMBP2 |
3 |
813 |
0.004192872 |
0.0011796520 |
1.829577 |
AAAAAA |
AAAAAA |
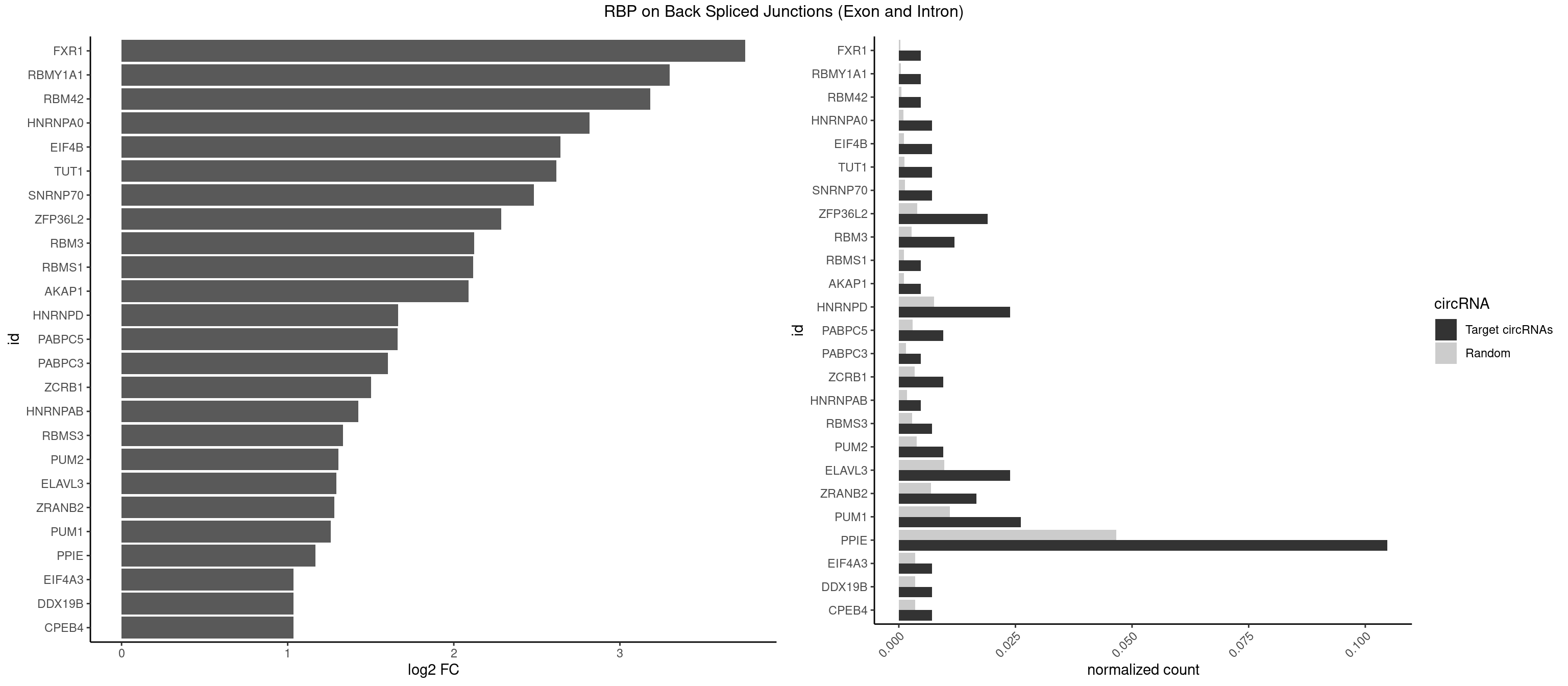
| FXR1 |
1 |
411 |
0.002096436 |
0.0005970720 |
1.811962 |
AUGACG |
ACGACA,ACGACG,AUGACA,AUGACG |
| PABPC4 |
5 |
1251 |
0.006289308 |
0.0018144033 |
1.793406 |
AAAAAA,AAAAAG |
AAAAAA,AAAAAG |
| RBFOX1 |
4 |
1077 |
0.005241090 |
0.0015622419 |
1.746249 |
AGCAUG,GCAUGC |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TRA2B |
32 |
7329 |
0.034591195 |
0.0106226650 |
1.703259 |
AAAGAA,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGGAAA,AGGAAG,GAAGAA,GAAUUA,GGAAGG |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| HNRNPA0 |
1 |
453 |
0.002096436 |
0.0006579386 |
1.671914 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| RBMX |
20 |
4925 |
0.022012579 |
0.0071387787 |
1.624579 |
AAGAAG,AAGGAA,AGGAAG,AUCAAA,AUCCCA,GGAAGG |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ESRP2 |
8 |
2150 |
0.009433962 |
0.0031172377 |
1.597596 |
GGGAAG,GGGGAA,GGGGAG,UGGGGA |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPN1 |
4 |
1222 |
0.005241090 |
0.0017723764 |
1.564182 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| HNRNPH3 |
30 |
7688 |
0.032494759 |
0.0111429292 |
1.544079 |
AGGGGA,GAAGGG,GGAAGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGAG |
AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| DAZAP1 |
23 |
5964 |
0.025157233 |
0.0086445016 |
1.541119 |
AAAAAA,AAUUUA,AGGAAA,AGGAAG,AGGAUG,UAGGAA,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPLL |
13 |
3534 |
0.014675052 |
0.0051229360 |
1.518323 |
ACACCA,ACCACA,CACACA,CACCAC,CAGACG,GCACAC |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPA1 |
67 |
18070 |
0.071278826 |
0.0261885646 |
1.444537 |
AAAAAA,AAAGAG,AAGAGG,AAUUUA,AGGAAG,AGGAGC,AUAGAA,AUUAGA,AUUUAA,CAAAGA,CAGGGA,GAGGAA,GAGGAG,GGAAGG,GGAGGA,GGCAGG,GGUGCG,UAGAGA,UAGGAA,UGGUGC,UUUUUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| RBM3 |
7 |
2152 |
0.008385744 |
0.0031201361 |
1.426330 |
AAAACG,AAGACG,GAAACU,GAUACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF7 |
52 |
14481 |
0.055555556 |
0.0209873716 |
1.404410 |
AAAGGA,AAGAAG,AAGGCG,AGAAGA,AGAGAA,AGAGAU,AGGAAG,AGGACA,AUAGAA,CAGAGA,CUGAGA,GAAGAA,GAAGGC,GAGAGA,GAGGAA,GAUUGA,GGAAGG,UAGAGA,UCAACA,UGAGAG,UGGACA |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| HNRNPH2 |
34 |
9711 |
0.036687631 |
0.0140746688 |
1.382193 |
AAGGCG,AAGGGG,AGGGGA,CUGGGG,GAAGGG,GGAAGG,GGAGGA,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGGAG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| LIN28A |
14 |
4315 |
0.015723270 |
0.0062547643 |
1.329874 |
AGGAGA,GGAGAA,GGAGGA |
AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF10 |
46 |
13860 |
0.049266247 |
0.0200874160 |
1.294308 |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGG,AGAGAA,AGAGGG,CAAAGA,GAGAAA,GAGAAG,GAGAGA,GAGAGG,GAGGAA,GAGGAG |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPAB |
5 |
1782 |
0.006289308 |
0.0025839306 |
1.283334 |
AAAGAC,AAGACA,CAAAGA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZCRB1 |
4 |
1605 |
0.005241090 |
0.0023274215 |
1.171134 |
AAUUAA,AUUUAA,GAAUUA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SYNCRIP |
4 |
1634 |
0.005241090 |
0.0023694485 |
1.145316 |
AAAAAA,UUUUUU |
AAAAAA,UUUUUU |
| SART3 |
7 |
2634 |
0.008385744 |
0.0038186524 |
1.134875 |
AAAAAA,AGAAAA,GAAAAA,GAAAAC |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SRSF6 |
65 |
21880 |
0.069182390 |
0.0317100317 |
1.125466 |
AAGAAG,AGAAGA,AGGAAG,AUCCUG,CACAGG,CACUGG,CAGCAC,CAUCCU,CCUCAG,CUCUGG,CUUCAG,GAAGAA,GAGGAA,GCAGCA,GGAAGA,UACAGG,UCACAC,UCUCAG,UGCGGA,UGCGUC,UGUGGA,UUACAG |
AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| HNRNPM |
2 |
999 |
0.003144654 |
0.0014492040 |
1.117641 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| SRSF11 |
1 |
688 |
0.002096436 |
0.0009985015 |
1.070102 |
AAGAAG |
AAGAAG |
| CELF4 |
4 |
1782 |
0.005241090 |
0.0025839306 |
1.020300 |
GGUGUG,GUGUGG |
GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |