circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000187079:+:11:12764178:12764434
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000187079:+:11:12764178:12764434 | ENSG00000187079 | ENST00000527636 | + | 11 | 12764179 | 12764434 | 256 | GUUUAUUUUCUUGAAAAGGCUCCAGGCUUCGGCUUGGAAAAUCCCACCGCCAAAAUUGAGCCCAGCAGCUGGAGCGGCAGUGAGAGCCCUGCCGAAAACAUGGAAAGGAUGAGUGACUCUGCAGAUAAGCCAAUUGACAAUGAUGCAGAAGGGGUCUGGAGCCCCGACAUCGAGCAAAGCUUUCAGGAGGCCCUGGCUAUCUAUCCACCAUGUGGGAGGAGGAAAAUCAUCUUAUCAGACGAAGGCAAAAUGUAUGGUUUAUUUUCUUGAAAAGGCUCCAGGCUUCGGCUUGGAAAAUCCCACCGC | circ |
| ENSG00000187079:+:11:12764178:12764434 | ENSG00000187079 | ENST00000527636 | + | 11 | 12764179 | 12764434 | 22 | CAAAAUGUAUGGUUUAUUUUCU | bsj |
| ENSG00000187079:+:11:12764178:12764434 | ENSG00000187079 | ENST00000527636 | + | 11 | 12763979 | 12764188 | 210 | CAAGCAAAGCUGGUGACUGAACCAAAAUCACCACUAAAGAACUUAACCGUGUAACCCAAAACCAUGUGUAUCCCCAAAAAGACUGAAAUAAUAAAAUGAAAUGAAAAAAGCUGGUGACUGAAGAACUUGCACUAGAGCUAGCAAGAGCAUUAUGUUUGUGGUUACCACAUCCUUAUACUGUUUUUGGUUUUCUCUUCUAGGUUUAUUUUC | ie_up |
| ENSG00000187079:+:11:12764178:12764434 | ENSG00000187079 | ENST00000527636 | + | 11 | 12764425 | 12764634 | 210 | AAAAUGUAUGGUAAGUGGCCUGGAACACUCCUUUGAAAUACUACAACCUGCAGUUGUGGUAGGGGAUAGAUUGUAGCUGGUCUUCCAGCAAGAGCUGUUCAUUGUAUGGGGUUGAGUGAUAUUUGUAGAAUUUUAGUGGGUCAUCCAAAGGACUCACCCUAAACUAAAAAGGAGAGUCAAUAAAGGACCUUUGGGAAAAAGAAAAGGACC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
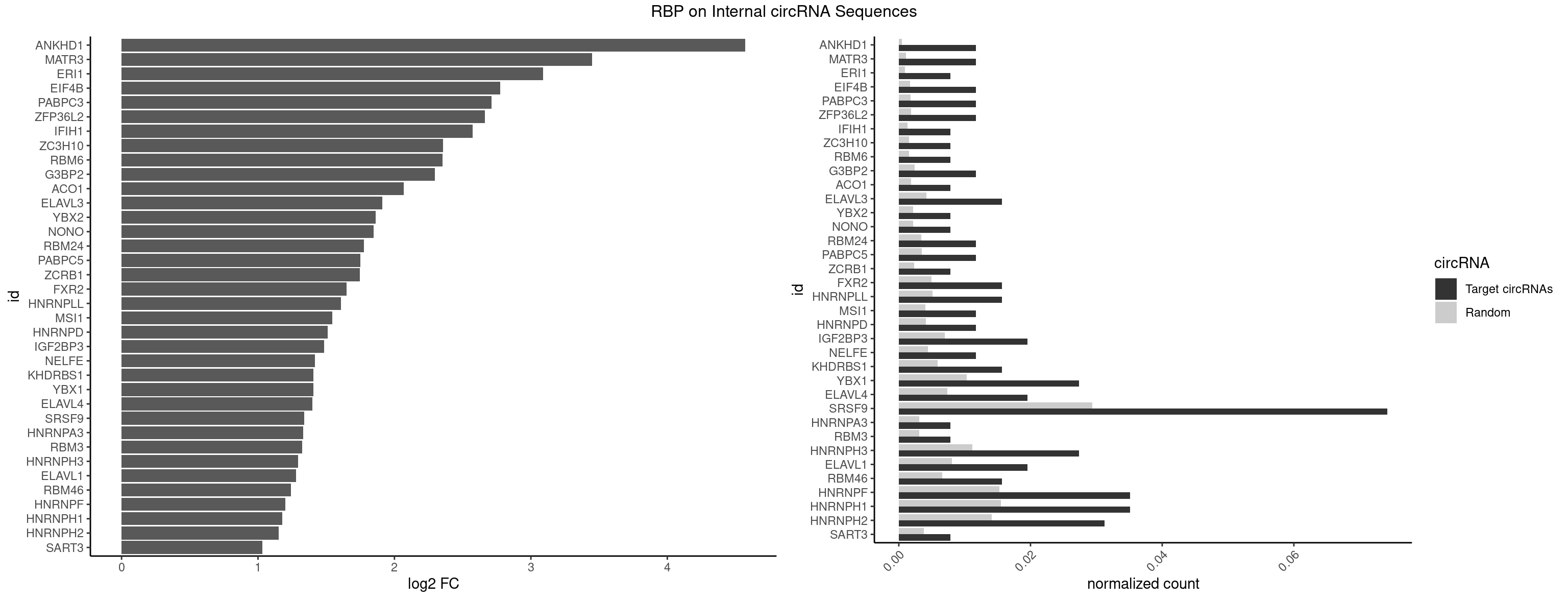
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 339 | 0.01171875 | 0.0004927293 | 4.571879 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| MATR3 | 2 | 739 | 0.01171875 | 0.0010724109 | 3.449889 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ERI1 | 1 | 632 | 0.00781250 | 0.0009173461 | 3.090246 | UUUCAG | UUCAGA,UUUCAG |
| EIF4B | 2 | 1179 | 0.01171875 | 0.0017100607 | 2.776699 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC3 | 2 | 1234 | 0.01171875 | 0.0017897669 | 2.710975 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZFP36L2 | 2 | 1277 | 0.01171875 | 0.0018520827 | 2.661598 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| IFIH1 | 1 | 904 | 0.00781250 | 0.0013115296 | 2.574534 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ZC3H10 | 1 | 1053 | 0.00781250 | 0.0015274610 | 2.354649 | GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM6 | 1 | 1054 | 0.00781250 | 0.0015289102 | 2.353281 | UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| G3BP2 | 2 | 1644 | 0.01171875 | 0.0023839405 | 2.297399 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ACO1 | 1 | 1283 | 0.00781250 | 0.0018607779 | 2.069878 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ELAVL3 | 3 | 2867 | 0.01562500 | 0.0041563169 | 1.910479 | UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| YBX2 | 1 | 1480 | 0.00781250 | 0.0021462711 | 1.863952 | ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 1 | 1498 | 0.00781250 | 0.0021723567 | 1.846523 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM24 | 2 | 2357 | 0.01171875 | 0.0034172229 | 1.777922 | GAGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| PABPC5 | 2 | 2400 | 0.01171875 | 0.0034795387 | 1.751851 | GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZCRB1 | 1 | 1605 | 0.00781250 | 0.0023274215 | 1.747052 | GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| FXR2 | 3 | 3434 | 0.01562500 | 0.0049780156 | 1.650214 | AGACGA,GACGAA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPLL | 3 | 3534 | 0.01562500 | 0.0051229360 | 1.608813 | AGACGA,CACCGC,CAGACG | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| MSI1 | 2 | 2770 | 0.01171875 | 0.0040157442 | 1.545079 | AGGAGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPD | 2 | 2837 | 0.01171875 | 0.0041128408 | 1.510612 | UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| IGF2BP3 | 4 | 4815 | 0.01953125 | 0.0069793662 | 1.484616 | AAAACA,AAAAUC,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| NELFE | 2 | 3028 | 0.01171875 | 0.0043896388 | 1.416645 | CUGGCU,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| KHDRBS1 | 3 | 4064 | 0.01562500 | 0.0058910141 | 1.407268 | CAAAAU,GAAAAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YBX1 | 6 | 7119 | 0.02734375 | 0.0103183321 | 1.406001 | ACAUCG,AUCAUC,CCACCA,CCAGCA,CCCUGC,GGUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ELAVL4 | 4 | 5105 | 0.01953125 | 0.0073996354 | 1.400258 | UAUCUA,UAUUUU,UUAUUU,UUUAUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| SRSF9 | 18 | 20254 | 0.07421875 | 0.0293536261 | 1.338245 | AGGAAA,GAAGGC,GAUGCA,GGAAAA,GGAAAG,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGGAGG,UGAAAA,UGACAA,UGAGAG,UGAUGC,UGGAGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| HNRNPA3 | 1 | 2140 | 0.00781250 | 0.0031027457 | 1.332239 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 1 | 2152 | 0.00781250 | 0.0031201361 | 1.324175 | AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPH3 | 6 | 7688 | 0.02734375 | 0.0111429292 | 1.295083 | AUGUGG,GAAGGG,GGAGGA,GGGAGG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| ELAVL1 | 4 | 5554 | 0.01953125 | 0.0080503280 | 1.278665 | UAUUUU,UGGUUU,UUAUUU,UUUAUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBM46 | 3 | 4554 | 0.01562500 | 0.0066011240 | 1.243073 | AAUCAU,AAUGAU,AUGAUG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPF | 8 | 10561 | 0.03515625 | 0.0153064921 | 1.199637 | AAGGGG,AUGUGG,GAAGGG,GAGGAA,GGAGGA,GGGAGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 8 | 10717 | 0.03515625 | 0.0155325680 | 1.178485 | AAGGGG,AUGUGG,GAAGGG,GAGGAA,GGAGGA,GGGAGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH2 | 7 | 9711 | 0.03125000 | 0.0140746688 | 1.150755 | AAGGGG,AUGUGG,GAAGGG,GGAGGA,GGGAGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| SART3 | 1 | 2634 | 0.00781250 | 0.0038186524 | 1.032721 | GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
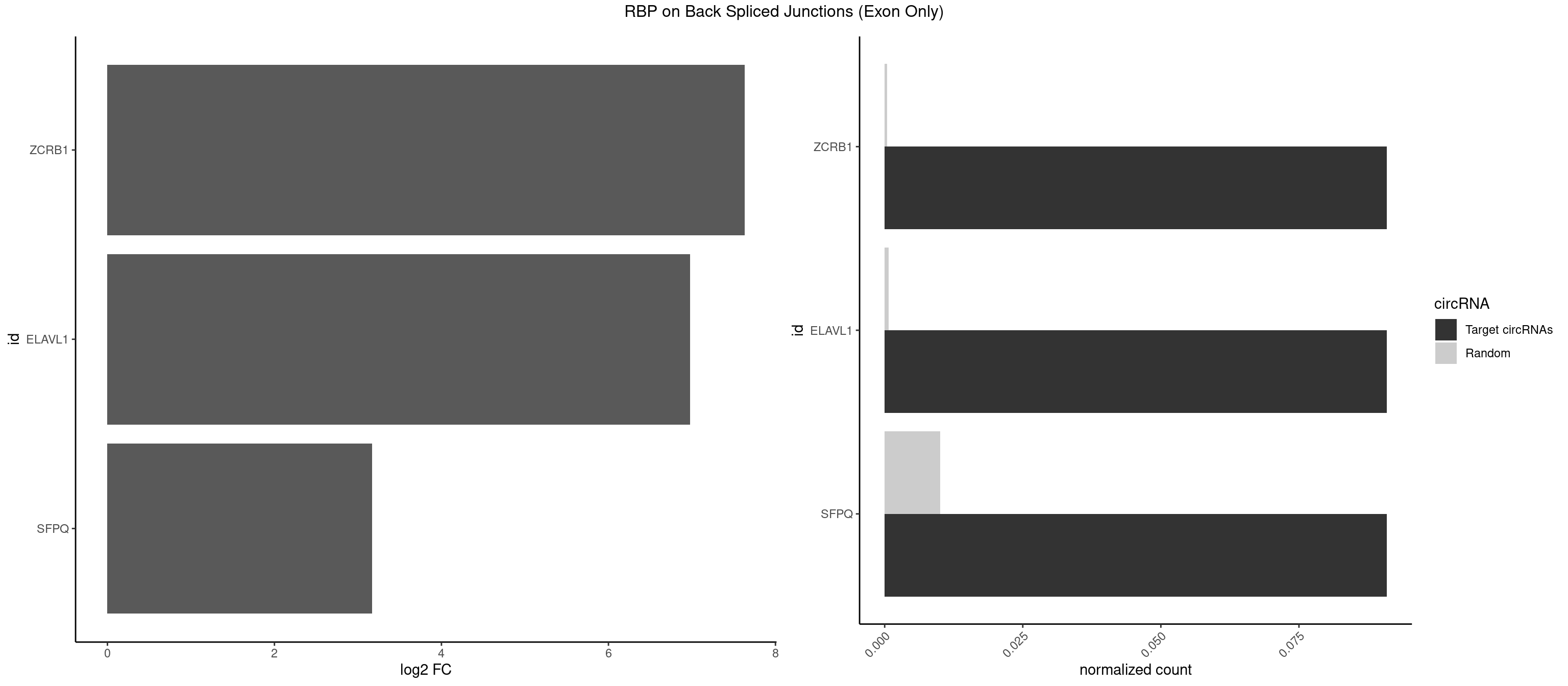
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZCRB1 | 1 | 6 | 0.09090909 | 0.0004584752 | 7.631437 | GGUUUA | AAUUAA,AUUUAA,GACUUA,GGCUUA |
| ELAVL1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UGGUUU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | UGGUUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
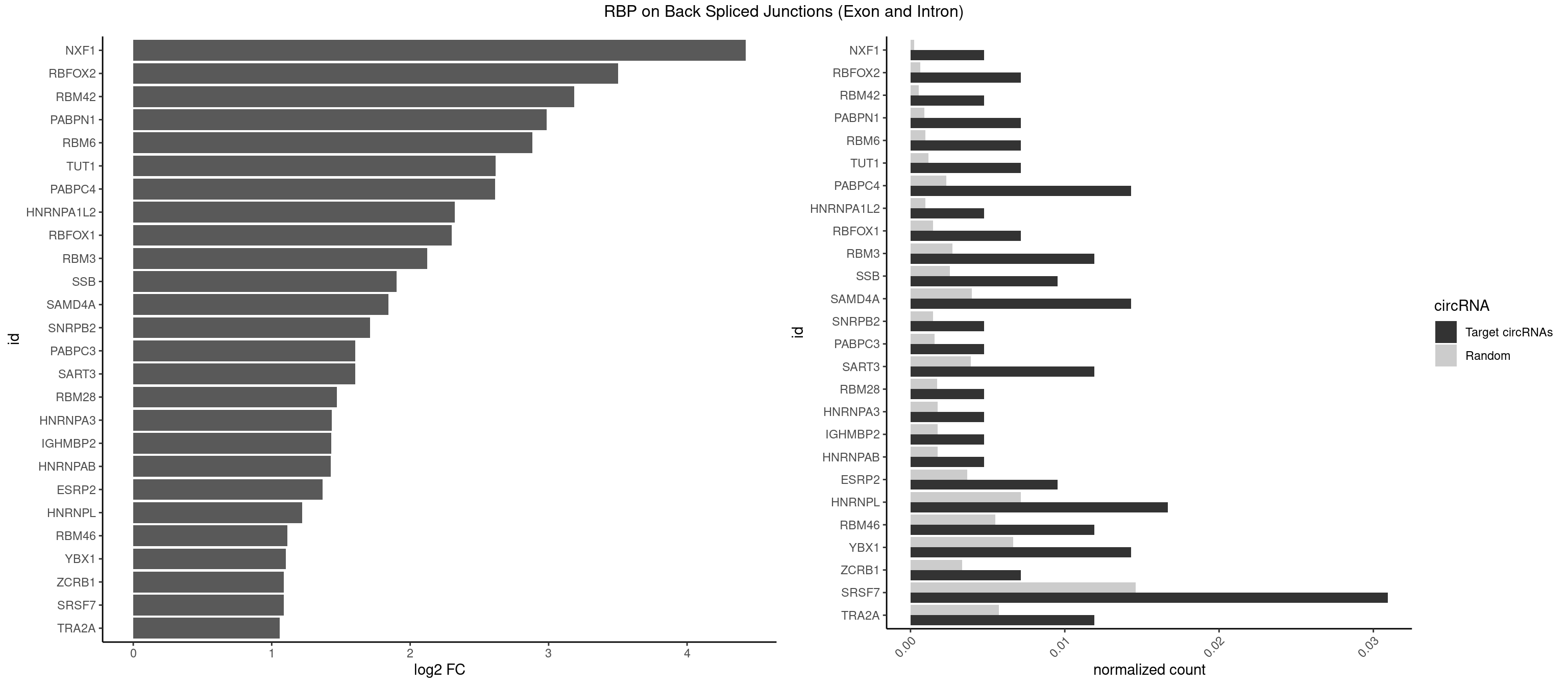
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGACUG | UGACUG,UGCAUG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA | AAAAGA,AGAAGA |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPC4 | 5 | 462 | 0.014285714 | 0.0023327287 | 2.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM3 | 4 | 541 | 0.011904762 | 0.0027307537 | 2.124168 | AAACUA,AAGACU,AAUACU,AUACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SAMD4A | 5 | 790 | 0.014285714 | 0.0039852882 | 1.841817 | CUGGAA,CUGGUC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SART3 | 4 | 777 | 0.011904762 | 0.0039197904 | 1.602690 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAAUAA,AAAUAC,AAUAAA,ACCACA,CACCAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUGAA,AUGAAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACCACA,CACCAC,CAGCAA,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | ACUUAA,GGUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAAGGA,AAGGAC,ACUAGA,CUAGAG,CUCUUC,GAAGAA,GAUAGA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAAGAA,AAGAAA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.