circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000172943:-:X:53984913:54002682
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000172943:-:X:53984913:54002682 | ENSG00000172943 | ENST00000686349 | - | X | 53984914 | 54002682 | 2085 | GGUGGAUCCAUGCUGUGCUGACGCCUGUGGACUGCCUUGCCUUUGGAGGGAACUUCUUACACAGCCUUAACAUCGAGAUGCAGCUCAAAGCCUAUGAGAUUGAGAAGCGGCUGAGCACAGCAGACCUCUUCAGAUUCCCCAACUUUGAGACCAUCUGUUGGUAUGUGGGAAAGCACAUCCUGGACAUCUUUCGCGGUUUGCGAGAGAACAGGAGACACCCUGCCUCCUACCUGGUCCAUGGUGGCAAAGCCUUGAACUUGGCCUUUAGAGCCUGGACAAGGAAAGAAGCUCUGCCAGACCAUGAGGAUGAGAUCCCGGAGACAGUGCGAACCGUACAGCUCAUUAAAGAUCUGGCCAGGGAGAUCCGCCUGGUGGAAGACAUCUUCCAACAGAACGUUGGGAAGACGAGCAAUAUCUUUGGGCUGCAGAGGAUCUUCCCAGCCGGCUCCAUUCCCCUAACCAGGCCAGCCCAUUCCACUUCAGUGUCCAUGUCCAGGCUGUCACUGCCCUCCAAAAAUGGUUCAAAGAAGAAAGGCCUGAAGCCCAAGGAACUCUUCAAGAAGGCAGAGCGAAAGGGCAAGGAGAGUUCAGCCUUGGGGCCUGCUGGCCAGUUGAGCUAUAAUCUCAUGGACACAUACAGUCAUCAGGCACUGAAGACAGGCUCUUUCCAGAAAGCAAAGUUCAACAUCACUGGUGCCUGCUUGAAUGACUCAGAUGACGACUCACCAGACUUGGACCUUGAUGGAAAUGAGAGCCCAUUGGCCCUAUUGAUGUCUAACGGCAGUACGAAAAGGGUGAAGAGUUUAUCCAAAUCUCGGCGAACCAAGAUAGCAAAGAAGGUAGACAAGGCUAGGCUGAUGGCAGAACAGGUGAUGGAAGACGAAUUUGACUUGGAUUCAGAUGAUGAGCUGCAGAUUGACGAGAGAUUGGGAAAGGAGAAGGCGACCCUGAUAAUAAGACCAAAAUUUCCCCGGAAAUUGCCCCGUGCGAAGCCUUGCUCUGACCCCAACCGAGUUCGUGAACCAGGAGAAGUUGAGUUUGACAUUGAGGAGGACUAUACAACAGAUGAGGACAUGGUGGAAGGGGUUGAAGGCAAGCUUGGGAAUGGUAGUGGCGCUGGUGGCAUUCUUGAUCUGCUCAAGGCCAGCAGGCAGGUGGGGGGACCUGACUAUGCUGCCCUCACGUGAGUACUGGCUUUUAUCUCCCCUUUCCAGGCUAUCAGACAGCAACCCCCGCUCCCGCCCAAGGUGCCAGGUCAGCCAAUCAGUAGAUAUUUAUAGAGCUAUUUAAAGGGACAAAGAAUUAUCCAGGGCAUUUGGGUGAAAGAAUAAAGUAGGCAUGAAACCACUCCUAAGGAACUUAAGAUCUGAUGGCAAUAAGAUGUAUAGACUUCAAGAGGAAUCAAUUUUGGGCACUUAGGAAGUUGGGAAGAGGGGCUGGAAGAGUCAAGCAAGGAGAAUUGUUACAAAAACAAUGAAAGGAAAGAAUGCAGCAUGCCUAAACAGGGAGGGAAGUGUGCCCCUUUUUUGCCACCUUGCCUGCUGUCUGUGGAGAGACAACACAAAAAGGACUGGCUUUUCGUGAAAGCAAUCUAUGAUACUGUCUUGAUCCUUAAACUUAAUUCCAAAUUUUUCACUUUGGAUGAUUCUCCCCUUUUUCCUCCCACUUUGAUCCUCAUAGCUAUCAUCCCUCCUCCCUGAGUAUUCUGAUGACCGAUUUACUCUGACAGCUAAAAACAACUCUCUCAGUAUUUCUCCUCUGUUCCUUGCAGCGAGGCCCCAGCUUCUCCCAGCACUCAGGAGGCCAUCCAGGGCAUGCUGUGCAUGGCCAACCUGCAGUCCUCAUCGUCCUCACCGGCUACCUCUAGCCUGCAGGCCUGGUGGACUGGGGGACAGGAUCGAAGCAGUGGGAGCUCCAGCAGUGGGCUGGGCACAGUGUCUAACAGUCCUGCUUCCCAGCGCACCCCAGGGAAGCGGCCCAUCAAGCGGCCAGCAUACUGGAGAACCGAGAGCGAGGAGGAGGAGGAGAACGCCAGUCUGGAUGAACAGGACAGCUUGGGAGCGUGCUUCAAGGAUGCAGAGUAUAGGUGGAUCCAUGCUGUGCUGACGCCUGUGGACUGCCUUGCCUUUGGAGGG | circ |
| ENSG00000172943:-:X:53984913:54002682 | ENSG00000172943 | ENST00000686349 | - | X | 53984914 | 54002682 | 22 | UGCAGAGUAUAGGUGGAUCCAU | bsj |
| ENSG00000172943:-:X:53984913:54002682 | ENSG00000172943 | ENST00000686349 | - | X | 54002673 | 54002882 | 210 | AGUGGCAUCUCAGCUUCUCUCAAAGAGUCUCAGGGAGCCUAAGGAUUCCUUGGACCACACCUUGAGAACAUCUGGUGUUGGGAAGCCAGUUUGGAGGCUAACUGCUCUUAAGGAGUUGGGUUUGAGAUGAGUAGGUAGUGGAGAUAAUCAGAGGAAGAAAGGCCCUCUUCCAGCUUUGAUUUUGCCUUGUUACACUGCAGGGUGGAUCCA | ie_up |
| ENSG00000172943:-:X:53984913:54002682 | ENSG00000172943 | ENST00000686349 | - | X | 53984714 | 53984923 | 210 | GCAGAGUAUAGUAAGGGCAGCUAAGCACAUGGGUUCAGGCCAGAAGGGGUCUCCUCAGUCCCAGUCUCCAUCCUAGUCCCAAUAGAGGACAGCUUGUAGUGAACAAAGUCCUGGUCUUAAGCCUGAUACUUAGUUUCUCUAAGCCAUAUUUUCCUAAUUUGUAAAAUGGGAAUAUCUCACAGGUUAAUGUUAAGAAUUUGAGAUUAUGCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
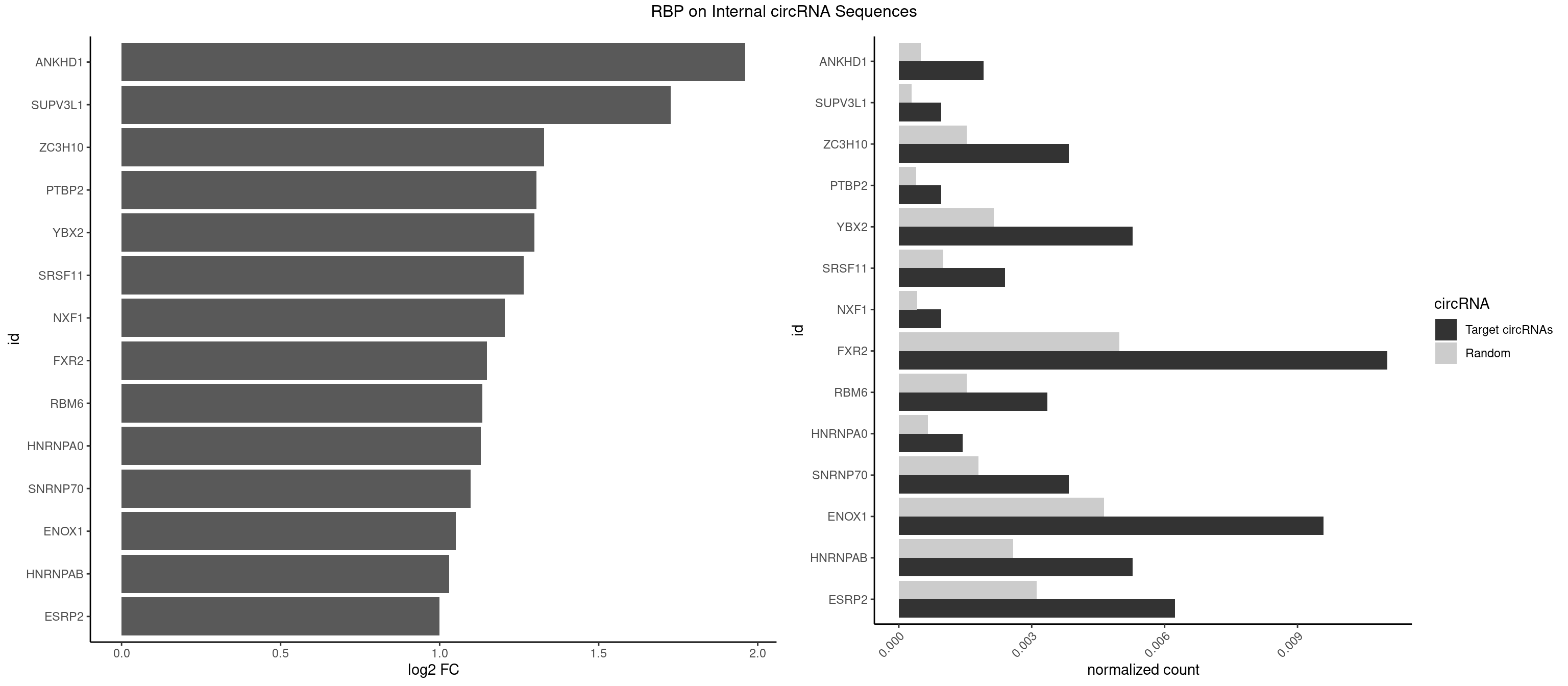
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 3 | 339 | 0.0019184652 | 0.0004927293 | 1.961085 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SUPV3L1 | 1 | 199 | 0.0009592326 | 0.0002898408 | 1.726620 | CCGCCC | CCGCCC |
| ZC3H10 | 7 | 1053 | 0.0038369305 | 0.0015274610 | 1.328817 | CAGCGA,CAGCGC,CCAGCG,GAGCGA,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PTBP2 | 1 | 267 | 0.0009592326 | 0.0003883867 | 1.304387 | CUCUCU | CUCUCU |
| YBX2 | 10 | 1480 | 0.0052757794 | 0.0021462711 | 1.297552 | AACAAC,AACAUC,ACAACA,ACAACU,ACAUCA,ACAUCG,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SRSF11 | 4 | 688 | 0.0023980815 | 0.0009985015 | 1.264044 | AAGAAG | AAGAAG |
| NXF1 | 1 | 286 | 0.0009592326 | 0.0004159215 | 1.205569 | AACCUG | AACCUG |
| FXR2 | 22 | 3434 | 0.0110311751 | 0.0049780156 | 1.147944 | AGACAA,AGACAG,AGACGA,GACAAA,GACAAG,GACAGG,GACGAA,GACGAG,GGACAA,GGACAG,UGACAG,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| RBM6 | 6 | 1054 | 0.0033573141 | 0.0015289102 | 1.134804 | AUCCAA,AUCCAG,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA0 | 2 | 453 | 0.0014388489 | 0.0006579386 | 1.128890 | AGAUAU,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| SNRNP70 | 7 | 1237 | 0.0038369305 | 0.0017941145 | 1.096681 | AAUCAA,AUCAAG,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ENOX1 | 19 | 3195 | 0.0095923261 | 0.0046316558 | 1.050353 | AAGACA,AGACAG,AGGACA,AUACAG,CAGACA,CAUACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPAB | 10 | 1782 | 0.0052757794 | 0.0025839306 | 1.029817 | AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ESRP2 | 12 | 2150 | 0.0062350120 | 0.0031172377 | 1.000124 | GGGAAA,GGGAAG,GGGAAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
RBP on BSJ (Exon Only)
Plot
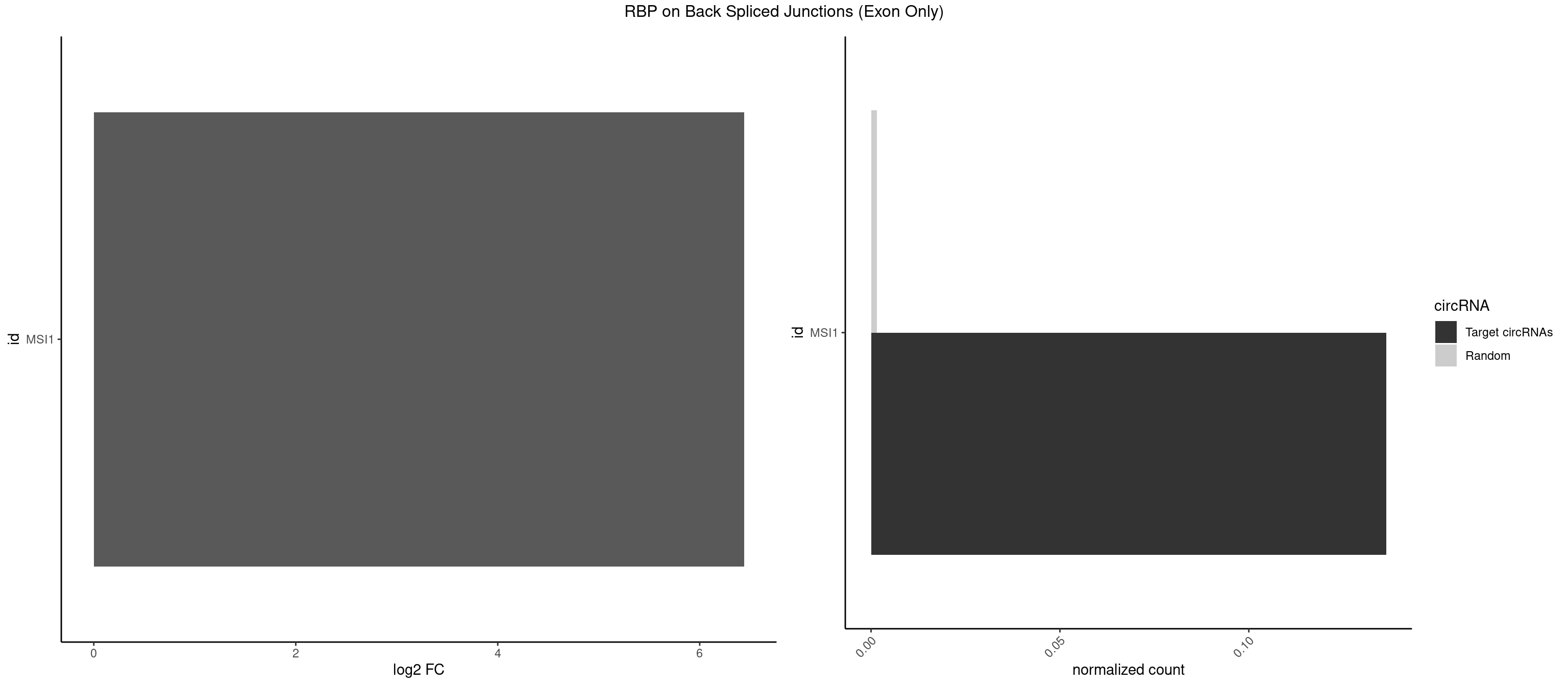
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MSI1 | 2 | 23 | 0.1363636 | 0.001571915 | 6.438792 | AGGUGG,UAGGUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUGG,UAGGAA,UAGGAG,UAGGUG,UAGUUA,UAGUUG |
RBP on BSJ (Exon and Intron)
Plot
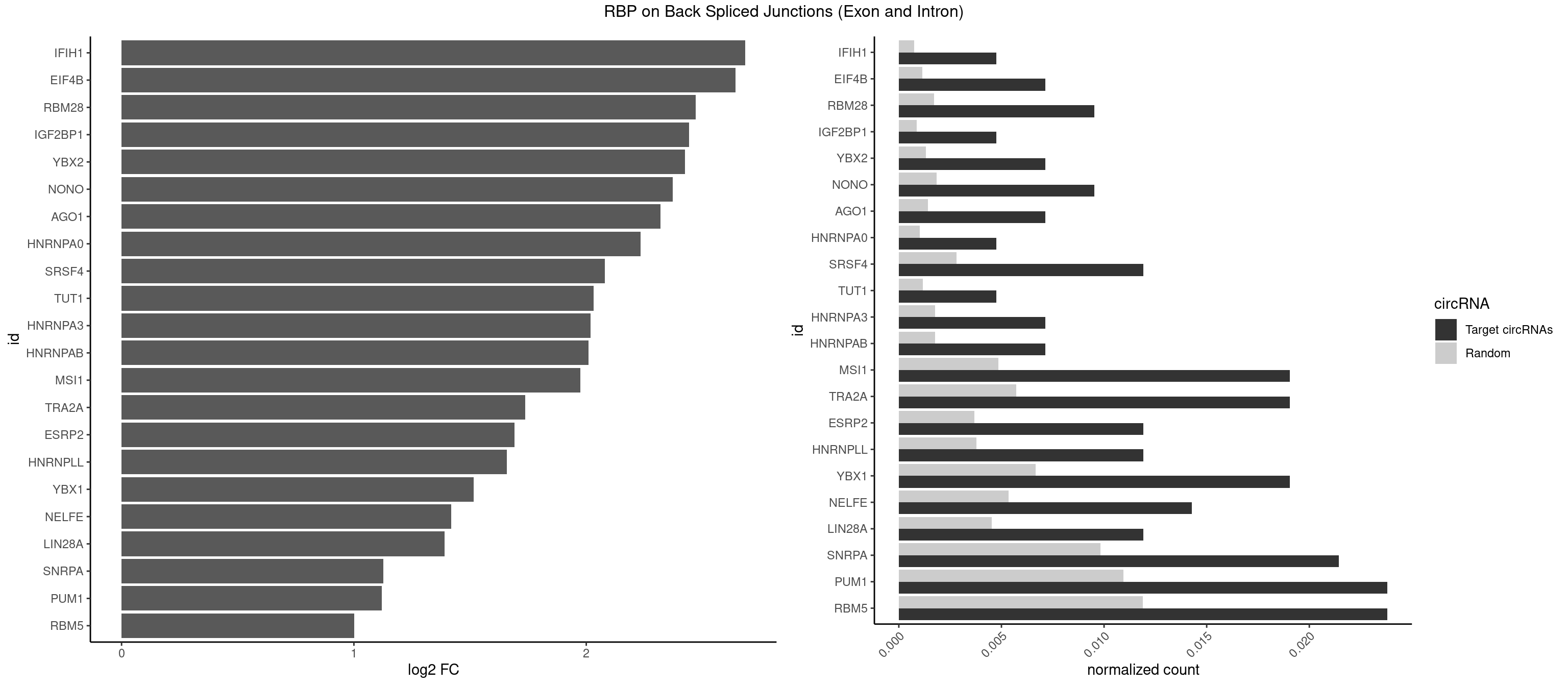
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 3 | 340 | 0.009523810 | 0.0017180572 | 2.470761 | AGUAGG,GAGUAG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAUC,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | AGAGGA,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| SRSF4 | 4 | 558 | 0.011904762 | 0.0028164047 | 2.079612 | AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MSI1 | 7 | 961 | 0.019047619 | 0.0048468360 | 1.974496 | AGGAAG,AGGUAG,AGUAAG,AGUAGG,AGUUGG,UAGGUA,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| TRA2A | 7 | 1133 | 0.019047619 | 0.0057134220 | 1.737184 | AAGAAA,AGAAAG,AGAGGA,AGGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ESRP2 | 4 | 731 | 0.011904762 | 0.0036880290 | 1.690617 | GGGAAG,GGGAAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACCACA,ACUGCA,CACACC,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| YBX1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | AACAUC,ACAUCU,ACCACA,CACACC,CAUCUG,CCACAC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | GCUAAC,GGCUAA,GGUCUC,UCUCUC,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGU,GGAGAU,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | AGUAGG,AUUCCU,GAUUCC,GGAGAU,GUAGUG,UAUGCU,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| PUM1 | 9 | 2172 | 0.023809524 | 0.0109482064 | 1.120844 | AAUGUU,AGAAUU,AGAUAA,CCAGAA,CUUGUA,UAAUGU,UGUAAA,UUAAUG,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AAGGAG,AAGGGG,AGGGAG,AGGGUG,CUCUUC,CUUCUC,GAAGGG,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.