circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000128710:+:2:176108906:176109358
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000128710:+:2:176108906:176109358 | ENSG00000128710 | MSTRG.19550.2 | + | 2 | 176108907 | 176109358 | 452 | UUGCCCCCCAGCGGUCCCGGAAAAAGCGCUGUCCCUAUACCAAGUACCAGAUCCGCGAACUGGAACGCGAGUUUUUCUUUAACGUGUACAUAAACAAAGAGAAAAGACUUCAACUCUCUCGGAUGCUCAACCUCACUGACCGGCAAGUCAAAAUCUGGUUCCAGAAUCGCAGGAUGAAAGAAAAGAAACUGAACAGAGACCGUCUGCAGUAUUUCACUGGAAACCCCUUAUUUUGAGAGCUCCAGGAAGCGCCCUCACCCCAGCCCCACUCACCCACCCUCCUUCCCACCAGCCUGCUCUCCGCAGGCCCACUGUCCUUGGGUUUAAUGACGUCUCUUCUCUGUGGAACUUCACGAUUCCUUCCCACGGUCAACUCGGGACCUCCCAGCGACCACUGCAGCCUGCGGACGAGGCCGGGACUUGGCCGAGCGGAUCCUAAUAAGGGGAAAAUGUUGCCCCCCAGCGGUCCCGGAAAAAGCGCUGUCCCUAUACCAAGUACCAG | circ |
| ENSG00000128710:+:2:176108906:176109358 | ENSG00000128710 | MSTRG.19550.2 | + | 2 | 176108907 | 176109358 | 22 | AGGGGAAAAUGUUGCCCCCCAG | bsj |
| ENSG00000128710:+:2:176108906:176109358 | ENSG00000128710 | MSTRG.19550.2 | + | 2 | 176108707 | 176108916 | 210 | GAACACAUGUCCACGCCCGCACUCUCUCCUGUGCCCGCCCAUAUAUCAUCCCCCACGACGCAGGCAGGGCCUUUCGGCCGCGGCAGAGAACGUCCCCAACGGGGCCAGGGCCUGGCCCUCGCUCAGUGGCCCGGGCGGGCGGGCGGGUGGGGGCUGUCAGGCAGCGGCCUCUCUCACCCCCUGGUCUCUUUGCCUUGCAGUUGCCCCCCA | ie_up |
| ENSG00000128710:+:2:176108906:176109358 | ENSG00000128710 | MSTRG.19550.2 | + | 2 | 176109349 | 176109558 | 210 | GGGGAAAAUGGUAAAUGCAAACGUCCCGUUACAAUUUUACCGCCAGUGUGCUGUCGUUCCCCCUCCCCCUCUCCGAGUCCUCGUGGGGACACGGCGGGGUCUGUAGGAAGUUGGGCCGGGUUGGGGGUUGCUAGAAGGCGCUGGUGUUUUGCUCUGAGUUUUAAGAGAUCCCUUCCUUCCUCUUCGGUGAAUGCAGGUUAUUUAAACUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
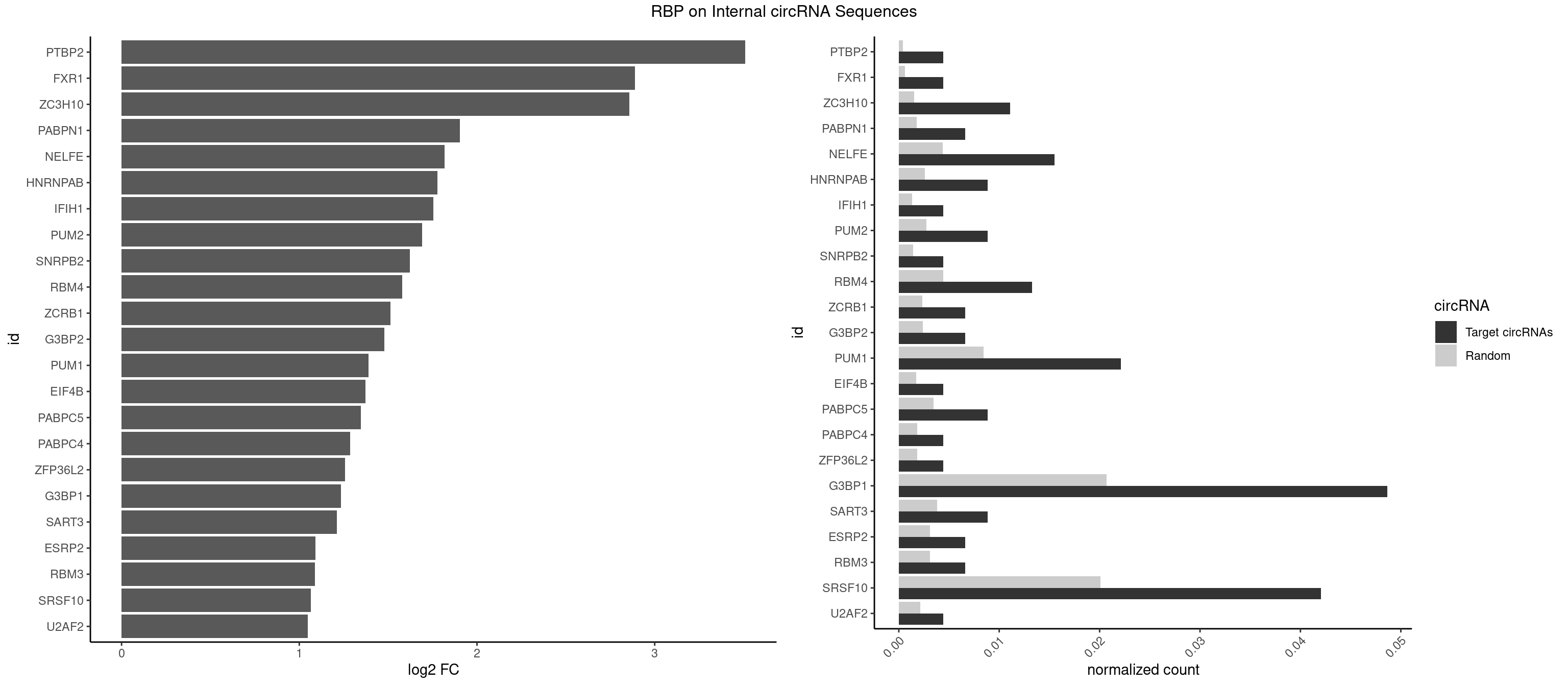
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.004424779 | 0.0003883867 | 3.510040 | CUCUCU | CUCUCU |
| FXR1 | 1 | 411 | 0.004424779 | 0.0005970720 | 2.889628 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| ZC3H10 | 4 | 1053 | 0.011061947 | 0.0015274610 | 2.856398 | CAGCGA,CCAGCG,CGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 2 | 1222 | 0.006637168 | 0.0017723764 | 1.904883 | AAAAGA | AAAAGA,AGAAGA |
| NELFE | 6 | 3028 | 0.015486726 | 0.0043896388 | 1.818858 | CUCUCU,CUGGUU,GUCUCU,UCUCUC,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| HNRNPAB | 3 | 1782 | 0.008849558 | 0.0025839306 | 1.776038 | AAAGAC,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| IFIH1 | 1 | 904 | 0.004424779 | 0.0013115296 | 1.754355 | GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| PUM2 | 3 | 1890 | 0.008849558 | 0.0027404447 | 1.691195 | GUACAU,UACAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SNRPB2 | 1 | 991 | 0.004424779 | 0.0014376103 | 1.621933 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM4 | 5 | 3065 | 0.013274336 | 0.0044432593 | 1.578949 | CCUUCC,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZCRB1 | 2 | 1605 | 0.006637168 | 0.0023274215 | 1.511835 | GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 2 | 1644 | 0.006637168 | 0.0023839405 | 1.477220 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| PUM1 | 9 | 5823 | 0.022123894 | 0.0084401638 | 1.390262 | AAUGUU,ACAUAA,CAGAAU,CCAGAA,GUACAU,UACAUA,UGUACA,UUAAUG,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| EIF4B | 1 | 1179 | 0.004424779 | 0.0017100607 | 1.371558 | CUCGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC5 | 3 | 2400 | 0.008849558 | 0.0034795387 | 1.346709 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPC4 | 1 | 1251 | 0.004424779 | 0.0018144033 | 1.286110 | AAAAAG | AAAAAA,AAAAAG |
| ZFP36L2 | 1 | 1277 | 0.004424779 | 0.0018520827 | 1.256457 | UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| G3BP1 | 21 | 14282 | 0.048672566 | 0.0206989801 | 1.233549 | ACCCAC,ACCCCU,ACCGGC,AGGCCC,AGGCCG,AUCCGC,CAGGCC,CCACCC,CCCACC,CCCACG,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCUCC,CCGCAG,CCGGCA,CUCCGC,UCCGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| SART3 | 3 | 2634 | 0.008849558 | 0.0038186524 | 1.212542 | AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ESRP2 | 2 | 2150 | 0.006637168 | 0.0031172377 | 1.090300 | GGGAAA,GGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM3 | 2 | 2152 | 0.006637168 | 0.0031201361 | 1.088959 | AAGACU,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SRSF10 | 18 | 13860 | 0.042035398 | 0.0200874160 | 1.065313 | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAGAAA,AAGAGA,AAGGGG,ACAAAG,AGAGAA,AGAGAC,CAAAGA,GAAAGA,GAGAAA,GAGACC,GAGAGC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| U2AF2 | 1 | 1477 | 0.004424779 | 0.0021419234 | 1.046698 | UUUUUC | UUUUCC,UUUUUC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
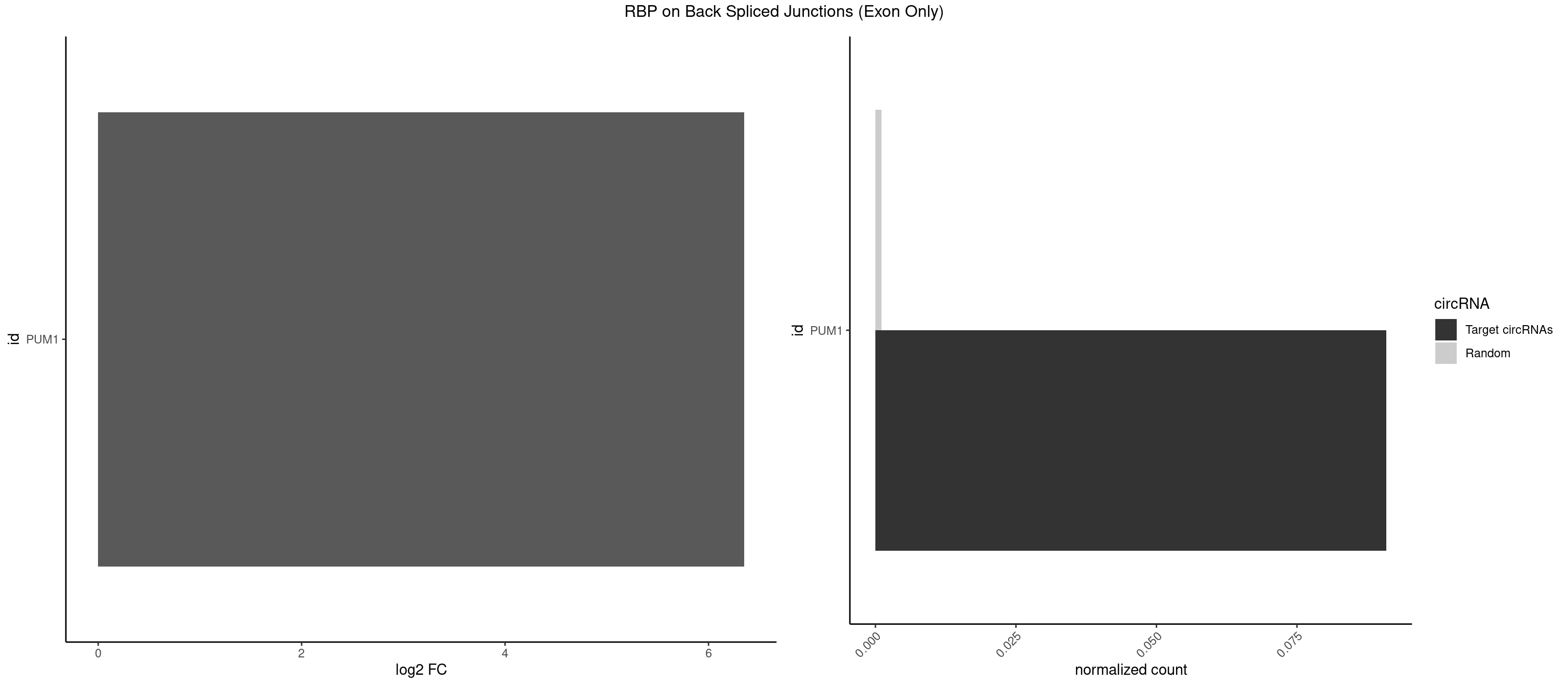
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PUM1 | 1 | 16 | 0.09090909 | 0.00111344 | 6.351329 | AAUGUU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
RBP on BSJ (Exon and Intron)
Plot
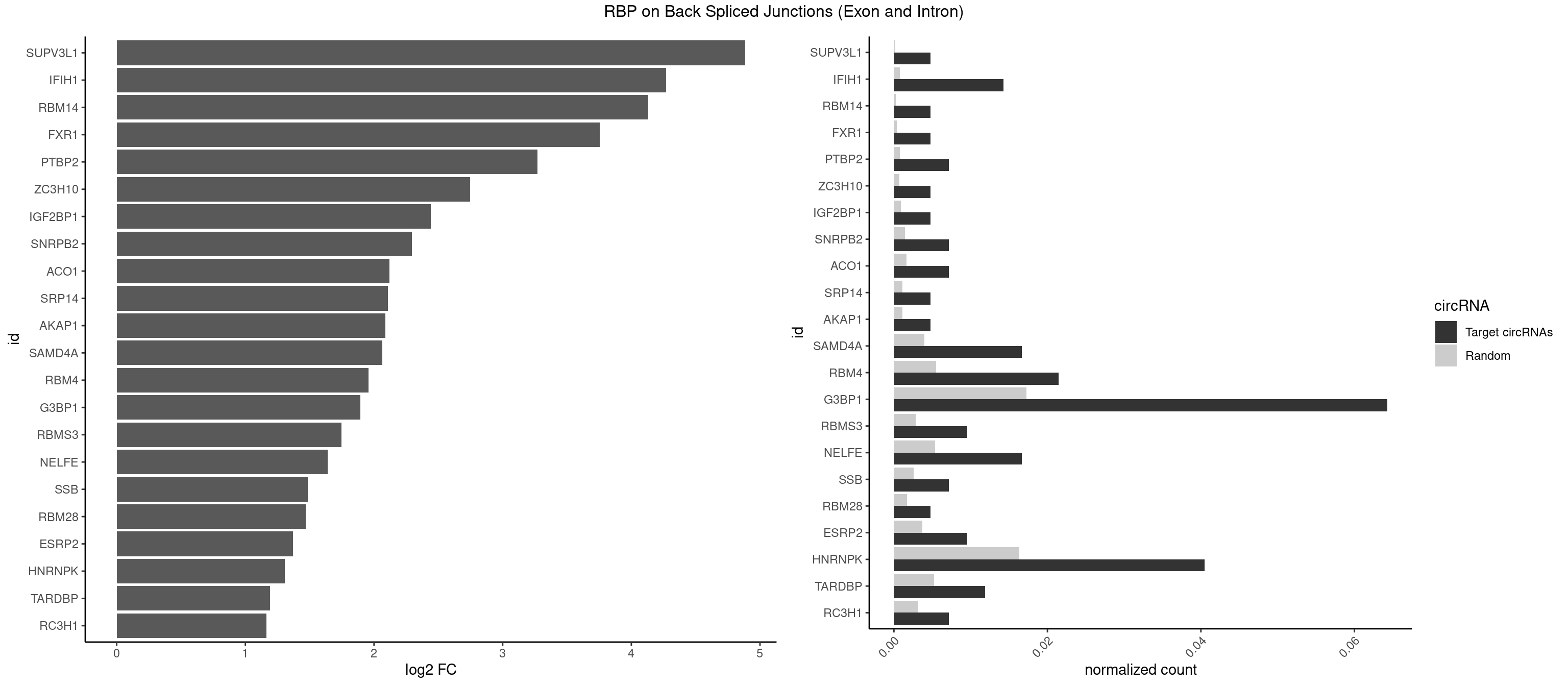
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 31 | 0.004761905 | 0.0001612253 | 4.884389 | CCGCCC | CCGCCC |
| IFIH1 | 5 | 146 | 0.014285714 | 0.0007406288 | 4.269679 | CCGCGG,GCCGCG,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM14 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGCGGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | ACGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| SAMD4A | 6 | 790 | 0.016666667 | 0.0039852882 | 2.064210 | CUGGCC,CUGGUC,GCGGGC,GCGGGU,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM4 | 8 | 1094 | 0.021428571 | 0.0055169287 | 1.957598 | CCUCUU,CCUUCC,CUCUUU,CUUCCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| G3BP1 | 26 | 3431 | 0.064285714 | 0.0172914148 | 1.894442 | ACCCCC,ACGCAG,ACGCCC,AGGCAG,CACGCC,CAGGCA,CCACGC,CCCACG,CCCAUA,CCCCAC,CCCCCA,CCCCCC,CCCCUC,CCCGCA,CCCGCC,CCCUCC,CCCUCG,CCGCAC,CCGCCC,CGGCAG,CGGCCG,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUCU,GGUCUC,GUCUCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPK | 16 | 3244 | 0.040476190 | 0.0163492543 | 1.307849 | ACCCCC,CAUCCC,CCCCAA,CCCCAC,CCCCCA,CCCCCC,CCCCCU,GCCCCC,UCCCCA,UCCCCC,UCCCUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGAAU,GUUUUG,UGAAUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCCUUC,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.