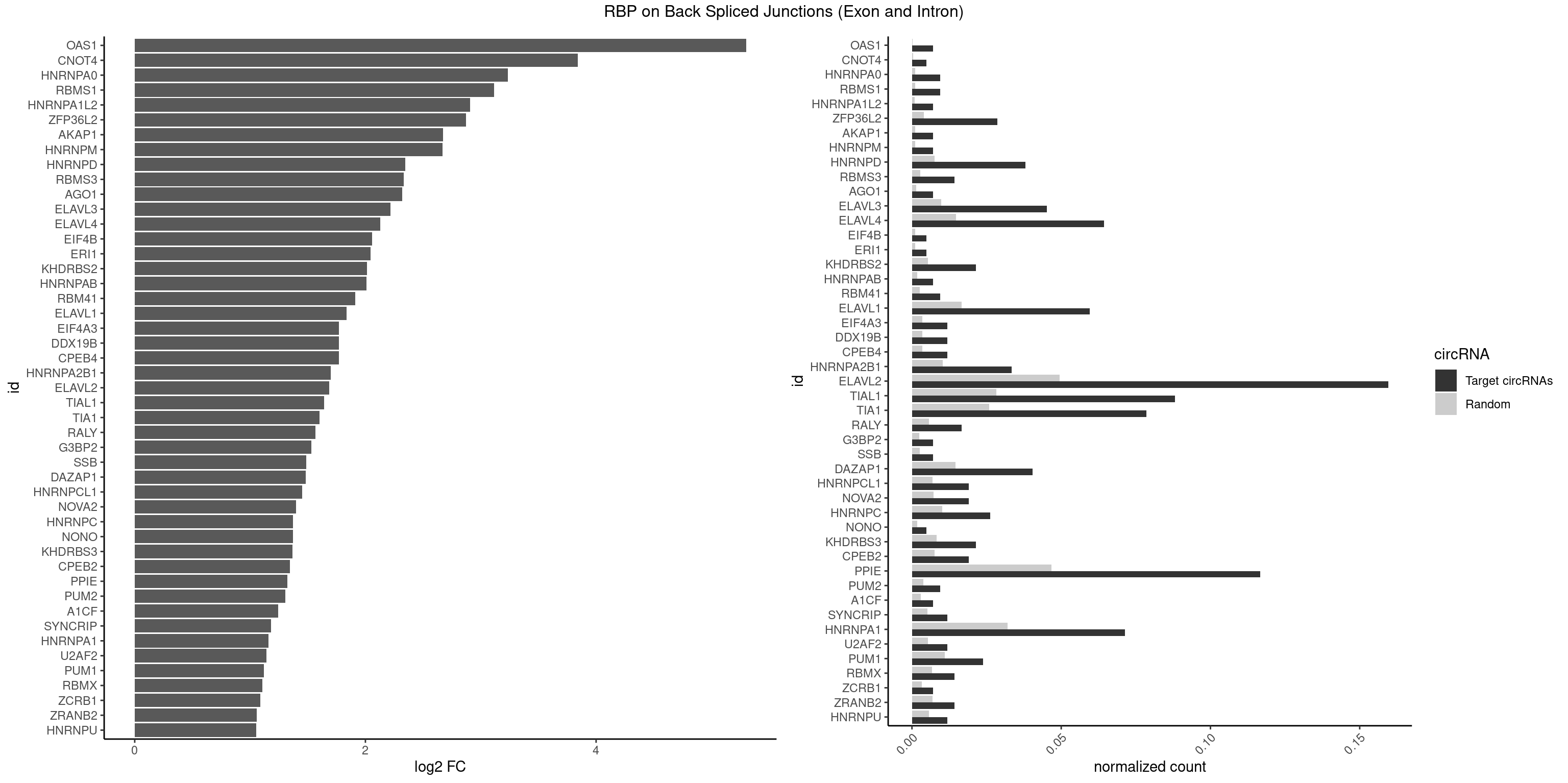
| OAS1 |
2 |
35 |
0.007142857 |
0.0001813785 |
5.299426 |
GCAUAA |
GCAUAA |
| CNOT4 |
1 |
65 |
0.004761905 |
0.0003325272 |
3.839994 |
GACAGA |
GACAGA |
| HNRNPA0 |
3 |
200 |
0.009523810 |
0.0010126965 |
3.233337 |
AAUUUA,AGAUAU |
AAUUUA,AGAUAU,AGUAGG |
| RBMS1 |
3 |
217 |
0.009523810 |
0.0010983474 |
3.116204 |
AUAUAG,GAUAUA,UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPA1L2 |
2 |
188 |
0.007142857 |
0.0009522370 |
2.907109 |
AUAGGG,UAGGGU |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ZFP36L2 |
11 |
774 |
0.028571429 |
0.0039046755 |
2.871299 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| AKAP1 |
2 |
221 |
0.007142857 |
0.0011185006 |
2.674935 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| HNRNPM |
2 |
222 |
0.007142857 |
0.0011235389 |
2.668451 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| HNRNPD |
15 |
1488 |
0.038095238 |
0.0075020153 |
2.344261 |
AAUUUA,AGAUAU,AUUUAU,UAUUUA,UUAGGA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBMS3 |
5 |
562 |
0.014285714 |
0.0028365578 |
2.332360 |
AUAUAG,AUAUAU,UAUAGC,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| AGO1 |
2 |
283 |
0.007142857 |
0.0014308746 |
2.319604 |
AGGUAG,GGUAGU |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ELAVL3 |
18 |
1928 |
0.045238095 |
0.0097188634 |
2.218679 |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ELAVL4 |
26 |
2916 |
0.064285714 |
0.0146966949 |
2.129006 |
AUUUAU,UAUCUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| EIF4B |
1 |
226 |
0.004761905 |
0.0011436921 |
2.057840 |
UUGGAA |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 |
1 |
228 |
0.004761905 |
0.0011537686 |
2.045185 |
UUCAGA |
UUCAGA,UUUCAG |
| KHDRBS2 |
8 |
1051 |
0.021428571 |
0.0053002821 |
2.015395 |
AAUAAA,AUAAAC,UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPAB |
2 |
351 |
0.007142857 |
0.0017734784 |
2.009919 |
AUAGCA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM41 |
3 |
502 |
0.009523810 |
0.0025342604 |
1.909974 |
UACUUG,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ELAVL1 |
24 |
3309 |
0.059523810 |
0.0166767432 |
1.835629 |
AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGUUUU,UUAUUU,UUGUUU,UUUAUU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| CPEB4 |
4 |
691 |
0.011904762 |
0.0034864974 |
1.771688 |
UUUUUU |
UUUUUU |
| DDX19B |
4 |
691 |
0.011904762 |
0.0034864974 |
1.771688 |
UUUUUU |
UUUUUU |
| EIF4A3 |
4 |
691 |
0.011904762 |
0.0034864974 |
1.771688 |
UUUUUU |
UUUUUU |
| HNRNPA2B1 |
13 |
2033 |
0.033333333 |
0.0102478839 |
1.701640 |
AAGGAA,AAUUUA,AGAUAU,AGGAAC,AUAGCA,AUAGGG,AUUUAA,UAGACA,UAGGGU,UUUAUA |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| ELAVL2 |
66 |
9826 |
0.159523810 |
0.0495112858 |
1.687942 |
AAUUUA,AAUUUU,AUUUAA,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUUA,GAUUUU,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAUACU,UAUAUA,UAUUAU,UAUUUA,UAUUUU,UCUUUU,UUAAGU,UUAAUU,UUACUU,UUAUAC,UUAUUU,UUCUUU,UUUAAG,UUUAAU,UUUAUA,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIAL1 |
36 |
5597 |
0.088095238 |
0.0282043531 |
1.643146 |
AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 |
32 |
5140 |
0.078571429 |
0.0259018541 |
1.600949 |
AUUUUC,AUUUUG,AUUUUU,CUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RALY |
6 |
1117 |
0.016666667 |
0.0056328094 |
1.565039 |
UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| G3BP2 |
2 |
490 |
0.007142857 |
0.0024738009 |
1.529772 |
AGGAUA,GGAUAA |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SSB |
2 |
505 |
0.007142857 |
0.0025493753 |
1.486358 |
UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| DAZAP1 |
16 |
2878 |
0.040476190 |
0.0145052398 |
1.480499 |
AAUUUA,AGAUAU,AGGAUA,AGGUAA,AGGUAG,AGUUUG,UAGGAU,UAGGUA,UAGGUU,UAGUUU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPCL1 |
7 |
1381 |
0.019047619 |
0.0069629182 |
1.451847 |
AUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| NOVA2 |
7 |
1434 |
0.019047619 |
0.0072299476 |
1.397554 |
AGUCAU,CUAGAU,GAUCAC,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPC |
10 |
2006 |
0.026190476 |
0.0101118501 |
1.372995 |
AUUUUU,UUUUUA,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| NONO |
1 |
364 |
0.004761905 |
0.0018389762 |
1.372636 |
AGGAAC |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| KHDRBS3 |
8 |
1646 |
0.021428571 |
0.0082980653 |
1.368689 |
AAUAAA,AUAAAC,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| CPEB2 |
7 |
1487 |
0.019047619 |
0.0074969770 |
1.345230 |
AUUUUU,CAUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PPIE |
48 |
9262 |
0.116666667 |
0.0466696896 |
1.321835 |
AAUAAA,AAUUUA,AAUUUU,AUAAUU,AUAUAU,AUUAUA,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAUAA,UAAUUU,UAUAUA,UAUUAU,UAUUUA,UAUUUU,UUAAUU,UUAUUA,UUAUUU,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| PUM2 |
3 |
764 |
0.009523810 |
0.0038542926 |
1.305073 |
UAGAUA,UAUAUA,UGUAAA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| A1CF |
2 |
598 |
0.007142857 |
0.0030179363 |
1.242939 |
AUAAUU,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SYNCRIP |
4 |
1041 |
0.011904762 |
0.0052498992 |
1.181177 |
UUUUUU |
AAAAAA,UUUUUU |
| HNRNPA1 |
29 |
6340 |
0.071428571 |
0.0319478033 |
1.160785 |
AAUUUA,AGAUAU,AGGGUU,AUAGCA,AUAGGG,AUUUAA,AUUUAU,GUAAGU,GUAGGU,UAGACA,UAGGAU,UAGGGU,UAGGUA,UUAUUU,UUUAUA,UUUAUU,UUUUUU |
AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| U2AF2 |
4 |
1071 |
0.011904762 |
0.0054010480 |
1.140228 |
UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| PUM1 |
9 |
2172 |
0.023809524 |
0.0109482064 |
1.120844 |
AGAAUU,CAGAAU,GUAAUA,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UUGUAC,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBMX |
5 |
1316 |
0.014285714 |
0.0066354293 |
1.106311 |
AAGGAA,AUCCCA,AUCCCC,UAAGAC |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZCRB1 |
2 |
666 |
0.007142857 |
0.0033605401 |
1.087808 |
ACUUAA,AUUUAA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| ZRANB2 |
5 |
1360 |
0.014285714 |
0.0068571141 |
1.058900 |
AAAGGU,AGGUAA,AGGUAG,AGGUUU |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPU |
4 |
1136 |
0.011904762 |
0.0057285369 |
1.055300 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |