circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000285733:-:6:169582835:169689012
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000285733:-:6:169582835:169689012 | ENSG00000285733 | MSTRG.29337.6 | - | 6 | 169582836 | 169689012 | 1643 | AUUGAAGAUGGAAAAUCCCCAAGACAUUUUCUCAAGUAAUGGUGGCUGUCUAAGUGAUAUAGUUAUAGAAAAAUACCUGGUUGAAUCCAAGGAGUCUGUGUCUCAUGUUCAGCUUGCUUGCAGCAUGCAGGACUGUGCUUUCCCUUUGGAUGGAACUGAACUUUGUAUAUGGAACACUAAGGAUCCUUCUCAUCAGCUUCUAAUCCUACGAGGACACCAUCAGCCAAUUACUGCUAUGGCUUUUGGAAAUAAAGUGAACCCACUUCUAAUCUGCUCAGCAUCACUGGAUUAUGUUAUAAUGUGGAACCUGGAUGAAUGUAGAGAGAAAGUACUUCAAGGGCUGGUCCCUCGAGGGACUGUCAUGGGCUCGCUUUUGGGAAAGGUGCUGUGUUUACAGUUGAGCCUGGAUGAUCAUGUUGUUGCCGUGUGUGCUGGAAACAAAAUAUUCAUGCUGGAUAUUGAGGUCUGGGACCAUUGUACAGGAUCAUUAAUAUACAGUUCAUCUGUGUUAUCAGCUUUGGAUCUUCAGUUUGAUGGAUGGACACCAUUAUCGUCGUGUGGCACGGGUUGACCUAAGGAAGAAGACAGAGACUUUCUCCACAAGAAGGGUUAAGUCUGGGCUGUGCAGCCAGCCAGAUGCCACAUGCACCCACCCAGCAACCUUGUCCUCACCUGGUCUCUGGCUGUCCAGAGUUGCUUCCUGUUGACGACAGCUCUCCUCAGAAGAAAGCCAGCUUCCCUCUACAAGUGCUCUGGGAAAAGGAGAGCAGGUUGAAGUAACAUUUCCUGUACUGAGACUUGCACCCUGUGAUCUCUCACUCAUCCCAAAUUCUGCAUGUGGAUGUCUUUCUUCUGAGAACACCCGAUGUGUGUGGAUCGGAAGCUCAGUGGGCUUAUUCGUAUUUAACCUGGCAAACCUGGAAGUGGAAGCUGCUUUGUAUUACAAGGAUUUCCAGAGCCUCAGCAUCCUGCUGGCCGGAUCGUGUGCCCUGAGGAACCGCACUGCGGAUCAAAAGGUGCUGUGCUUGCUGGCCUCCCUCUUUGGCGGGAAGAUUGCCGUGUUGGAGAUCAACCCGGCCGCGCUAGUCAGGGCUCAGCAGUGCCCCAGCAUGGGGCAGAGCCUCUCGGUGCCAGCCAGCUCCUGUGUCCUACCGACCUCUCCACUGUAUCUGGGAAUUGCCAAGGAGAAGAGUACCAAGGCUGCUAGUGAACAGCGACGUGAUAUAAACAGAAGAGCAAGUCCAAGCUGAUUUGCAGGCUCUCCACGACGGGUGCAGUAGACAUGACCAGUUUAUCGGCAGUCAACGACUUUUAUUCCCACAUCGUACUCGCAGCUGGCCGGAACAGGACCGUGGAAGUGUUUGACCUCAACGCCGGCUGCAGUGCAGCGGUGAUAGCGGAAGCCCACUCACGGCCUGUCCAUCAAAUCUGCCAAAAUAAAGUGUGAGCGCCACUUUGAAGGGCAUCCAACCCGCGGCUAUCCAUGUGGAAUCGCUUUCAGUCCUUGUGGACGAUUCGCGGCUUGUGGGGCCGAGGACAGACACGCUUACGUGUAUGAAAUGGGCUCAAGCACGUUUUCUCACCGGCUGGCUGGGCACACAGACACUGUCACUGGAGUGGCCUUCAACCCAUCAGCGCCCCAGAUUGAAGAUGGAAAAUCCCCAAGACAUUUUCUCAAGUAAUGGUGGCUGUC | circ |
| ENSG00000285733:-:6:169582835:169689012 | ENSG00000285733 | MSTRG.29337.6 | - | 6 | 169582836 | 169689012 | 22 | CAGCGCCCCAGAUUGAAGAUGG | bsj |
| ENSG00000285733:-:6:169582835:169689012 | ENSG00000285733 | MSTRG.29337.6 | - | 6 | 169689003 | 169689212 | 210 | AAUGGAGCAGUUGAGAAUGUGGAAGCUCCAGAGUUUUCGGCAAUUUCUCUUCUUUUUUCUUGGGAUGAGGAGAGGUGAAUAAAUAGGUAUGAUGUGGUAUAUGUGUGUGGUCAAACUAGUAACUAUCAGUAGGUAAUAGACUUUUUUUCUGUAUUAAAUAUGAUACCUAUAGUCAUCUUAUAUGUACUUUUUGUUUUCAGAUUGAAGAUG | ie_up |
| ENSG00000285733:-:6:169582835:169689012 | ENSG00000285733 | MSTRG.29337.6 | - | 6 | 169582636 | 169582845 | 210 | AGCGCCCCAGGUACAGUCUUGCUGAGUGGGUGGGGCGCCUCUGCUGCAUACAAGUCUUACAUUAUAUUUUUGUAUCUUUAUGACUUUAUUUUGGUGAAAUCUUAUAAUACUUAAGGUUACAUGCCAGUGUUCACUGAGUGCAAGGAGAACUUAGGAAAUAUAUUUUUAUUGGCGAGAGAGAGAGAAAUAAACCUUUAUUGUGGAGAAAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
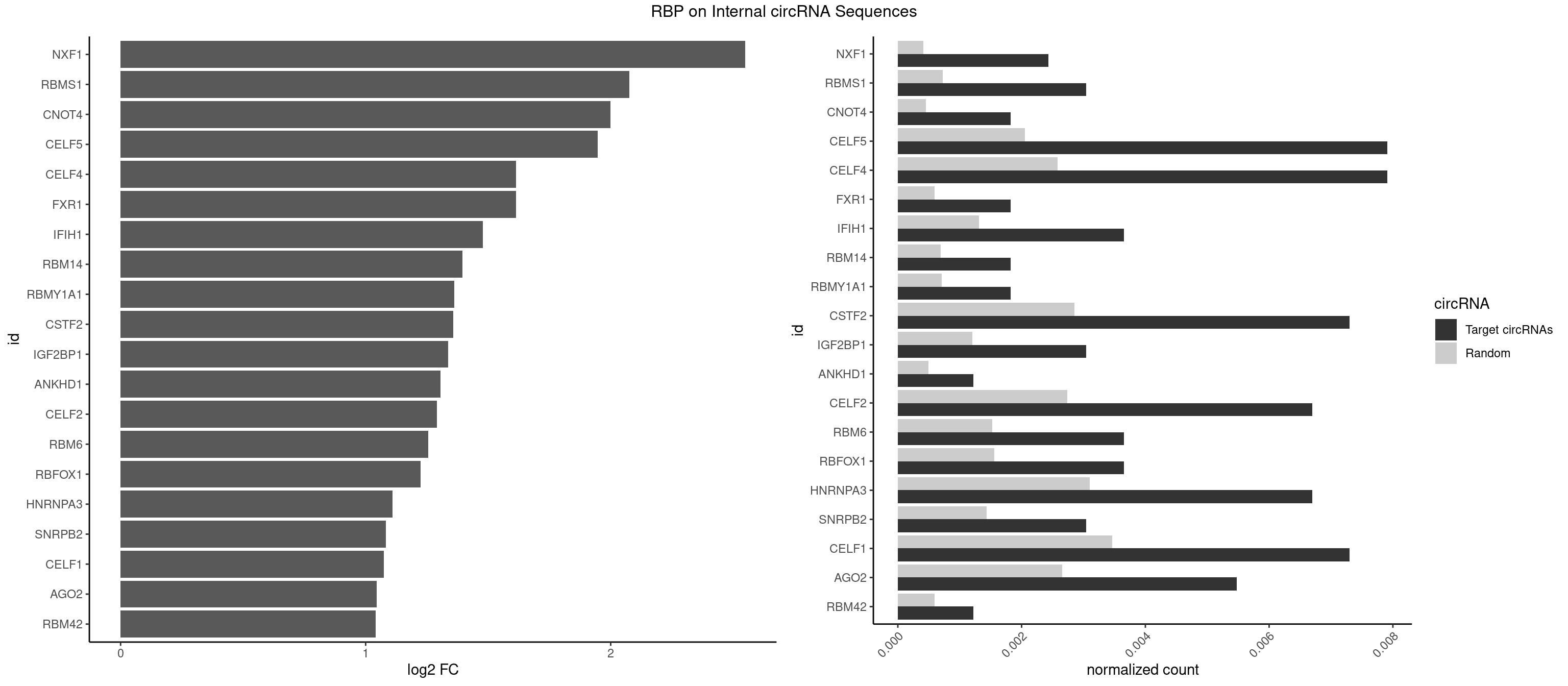
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 3 | 286 | 0.002434571 | 0.0004159215 | 2.549284 | AACCUG | AACCUG |
| RBMS1 | 4 | 497 | 0.003043214 | 0.0007217036 | 2.076117 | AUAUAC,AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| CNOT4 | 2 | 314 | 0.001825928 | 0.0004564992 | 1.999946 | GACAGA | GACAGA |
| CELF5 | 12 | 1415 | 0.007912355 | 0.0020520728 | 1.947025 | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 12 | 1782 | 0.007912355 | 0.0025839306 | 1.614540 | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| FXR1 | 2 | 411 | 0.001825928 | 0.0005970720 | 1.612653 | ACGACA,ACGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| IFIH1 | 5 | 904 | 0.003651856 | 0.0013115296 | 1.477380 | CCGCGG,GCCGCG,GCGGAU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM14 | 2 | 478 | 0.001825928 | 0.0006941687 | 1.395272 | CGCGGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBMY1A1 | 2 | 489 | 0.001825928 | 0.0007101099 | 1.362516 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| CSTF2 | 11 | 1967 | 0.007303713 | 0.0028520334 | 1.356639 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| IGF2BP1 | 4 | 831 | 0.003043214 | 0.0012057377 | 1.335680 | AAGCAC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ANKHD1 | 1 | 339 | 0.001217285 | 0.0004927293 | 1.304800 | GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| CELF2 | 10 | 1886 | 0.006695070 | 0.0027346479 | 1.291744 | AUGUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM6 | 5 | 1054 | 0.003651856 | 0.0015289102 | 1.256126 | AAUCCA,AUCCAA,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBFOX1 | 5 | 1077 | 0.003651856 | 0.0015622419 | 1.225012 | AGCAUG,GCAUGC,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA3 | 10 | 2140 | 0.006695070 | 0.0031027457 | 1.109554 | AAGGAG,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SNRPB2 | 4 | 991 | 0.003043214 | 0.0014376103 | 1.081923 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF1 | 11 | 2391 | 0.007303713 | 0.0034664959 | 1.075152 | CUGUCU,GUGUGU,GUGUUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| AGO2 | 8 | 1830 | 0.005477785 | 0.0026534924 | 1.045700 | AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| RBM42 | 1 | 407 | 0.001217285 | 0.0005912752 | 1.041766 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
RBP on BSJ (Exon Only)
Plot
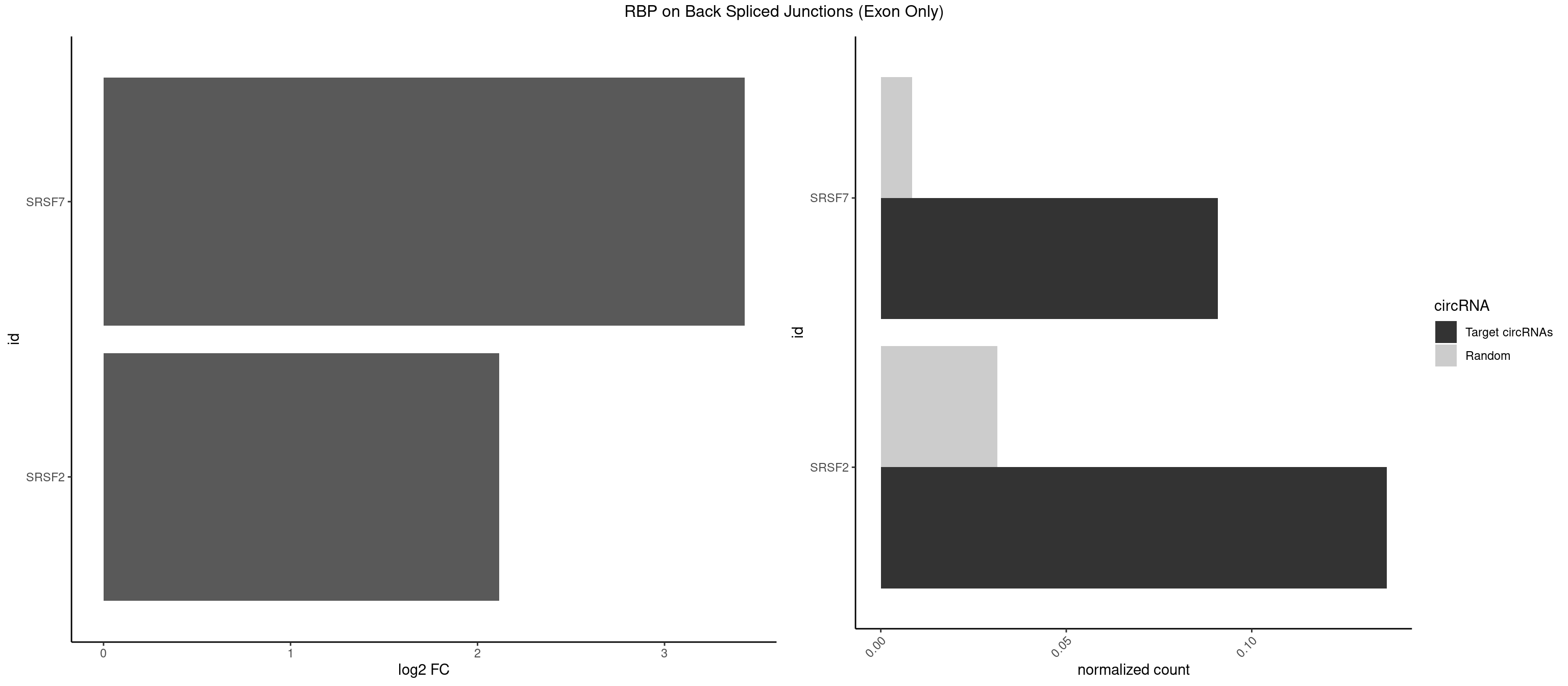
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF7 | 1 | 128 | 0.09090909 | 0.008449044 | 3.427565 | GAUUGA | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
| SRSF2 | 2 | 479 | 0.13636364 | 0.031438302 | 2.116864 | CCAGAU,GAUUGA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
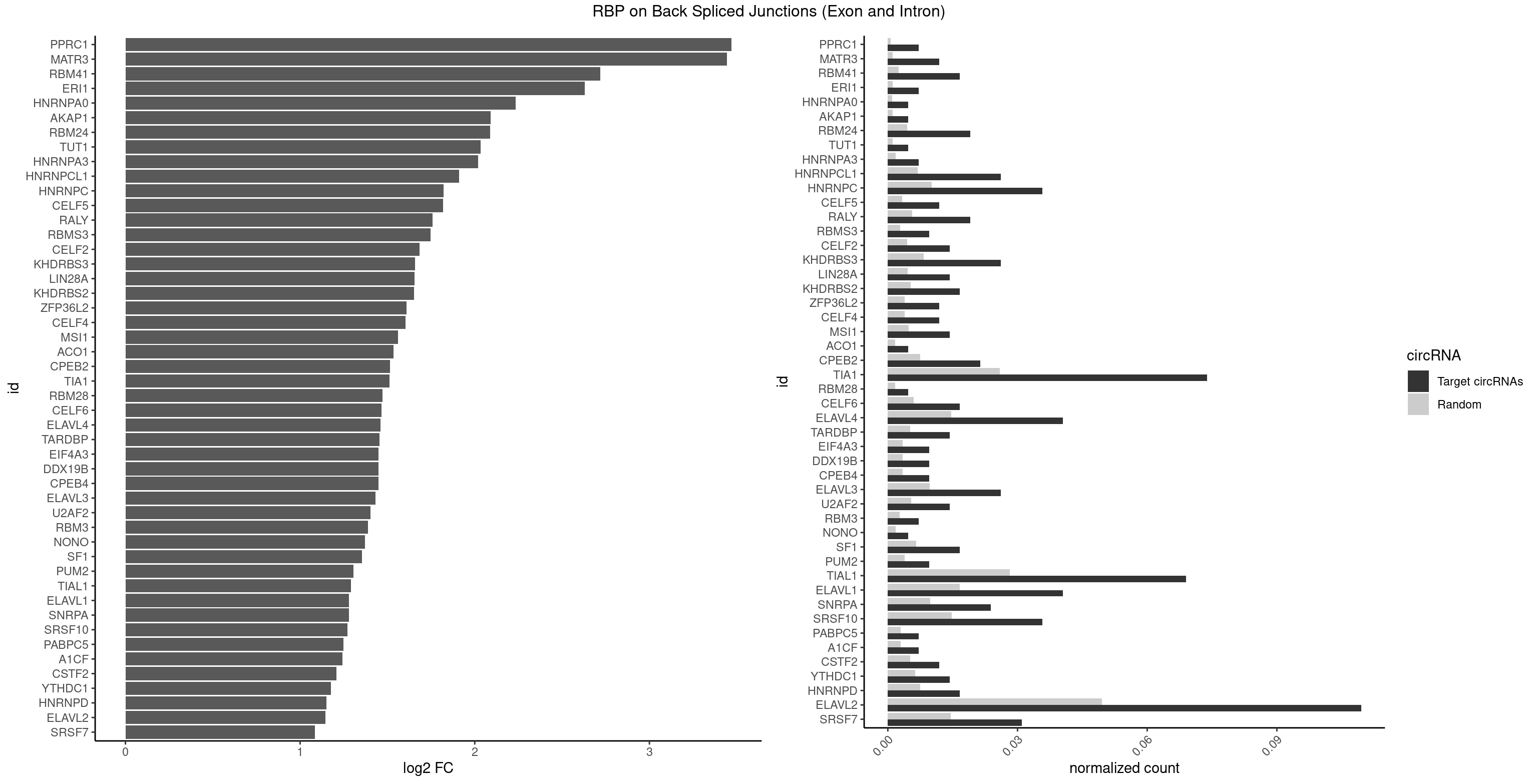
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| MATR3 | 4 | 216 | 0.011904762 | 0.0010933091 | 3.444765 | AAUCUU,AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM41 | 6 | 502 | 0.016666667 | 0.0025342604 | 2.717329 | AUACUU,UACAUG,UACAUU,UACUUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBM24 | 7 | 888 | 0.019047619 | 0.0044790407 | 2.088349 | GAGUGG,GUGUGG,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPCL1 | 10 | 1381 | 0.026190476 | 0.0069629182 | 1.911278 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPC | 14 | 2006 | 0.035714286 | 0.0101118501 | 1.820454 | AUUUUU,CUUUUU,GCAUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGG,GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RALY | 7 | 1117 | 0.019047619 | 0.0056328094 | 1.757684 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AAUAUA,AUAUAU,CUAUAG | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,GUGUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| KHDRBS3 | 10 | 1646 | 0.026190476 | 0.0082980653 | 1.658195 | AAAUAA,AAUAAA,AUAAAC,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | AGGAGA,GGAGAA,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| KHDRBS2 | 6 | 1051 | 0.016666667 | 0.0053002821 | 1.652825 | AAUAAA,AUAAAC,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGUAGG,UAGGAA,UAGGUA,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| TIA1 | 30 | 5140 | 0.073809524 | 0.0259018541 | 1.510752 | AUUUUG,AUUUUU,CUUUUU,GUUUUC,UAUUUU,UUAUUU,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CELF6 | 6 | 1196 | 0.016666667 | 0.0060308343 | 1.466536 | GUGGGG,GUGUGG,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| ELAVL4 | 16 | 2916 | 0.040476190 | 0.0146966949 | 1.461582 | GUAUCU,UAUUUU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGU,GUGAAU,GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| ELAVL3 | 10 | 1928 | 0.026190476 | 0.0097188634 | 1.430183 | UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| U2AF2 | 5 | 1071 | 0.014285714 | 0.0054010480 | 1.403262 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAACUA,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACAGUC,AGUAAC,CACUGA,UAGUAA,UGCUGA,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUAUAU,UAAAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TIAL1 | 28 | 5597 | 0.069047619 | 0.0282043531 | 1.291674 | AGUUUU,AUUUUG,AUUUUU,CUUUUU,GUUUUC,UAUUUU,UUAAAU,UUAUUU,UUUAUU,UUUCAG,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | UAUUUU,UGUUUU,UUAUUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| SNRPA | 9 | 1947 | 0.023809524 | 0.0098145909 | 1.278539 | AGCAGU,AGGAGA,AGUAGG,AUACCU,CAGUAG,GAGCAG,GAUACC,GGUAUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SRSF10 | 14 | 2937 | 0.035714286 | 0.0148024990 | 1.270661 | AAGGAG,AGAGAA,AGAGAG,GAGAAA,GAGAAC,GAGAGA,GAGAGG,GAGGAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUCAGU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAGUGC,GCAUAC,UAAUAC,UGAUAC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AGUAGG,UUAGGA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL2 | 45 | 9826 | 0.109523810 | 0.0495112858 | 1.145415 | AUACUU,AUAUUU,AUUUUG,AUUUUU,CUUAUA,CUUUUU,GUUUUC,UACUUU,UAUAUU,UAUUGU,UAUUUU,UCUUAU,UCUUUU,UGUAUU,UUAUAU,UUAUUG,UUAUUU,UUCUUU,UUGUAU,UUUAUG,UUUAUU,UUUCUU,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AGAGAA,AGAGAG,AUAGAC,CGAGAG,CUCUUC,GAGAGA,GAUUGA | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.