circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000188352:+:9:20862577:20867012
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000188352:+:9:20862577:20867012 | ENSG00000188352 | ENST00000380249 | + | 9 | 20862578 | 20867012 | 270 | GUUGUUUGCAUUCGCUCCACUUGGAAUGCUCUCUCUCCAAAGCUGAGUUGUGACACAAGACCUCUCAUUCUGAAGACACUGAGUGAACUAUUUUCUCUAGUUCCUUCCUUAACGGUCAAUACAACUGAAUAUGAGAAUUUUAAAGUUCAAGUCCUCAGCUUCCUCUGGACUCAUACUCAAAACAAGGACCCAAUUGUAGCAAAUGCUGCAUAUAGAUCCCUGGCCAACUUUAGUGCAGGAGAACACACCAUUCUUCAUCUGCCUGAAAAGGUUGUUUGCAUUCGCUCCACUUGGAAUGCUCUCUCUCCAAAGCUGAGUUG | circ |
| ENSG00000188352:+:9:20862577:20867012 | ENSG00000188352 | ENST00000380249 | + | 9 | 20862578 | 20867012 | 22 | UGCCUGAAAAGGUUGUUUGCAU | bsj |
| ENSG00000188352:+:9:20862577:20867012 | ENSG00000188352 | ENST00000380249 | + | 9 | 20862378 | 20862587 | 210 | GUGAGGCAGGCAUAACUUCAACAGUUUACACUGGUUUAAGUUAUUUCAUUUGAGGGGUGGUACCUUAUUAAGUCCUAUCUUCUAGGCAUAGUCUUAUUUGUUUCUUAAGUUUCCUUAUAAUAAAAUAUAGUACUCUUAAUUAAUGGCUUCAUAUAGGUUGGAGUCUUGUUAGGUGUUGACCUUUUCUAUUUGCUUCACAGGUUGUUUGCA | ie_up |
| ENSG00000188352:+:9:20862577:20867012 | ENSG00000188352 | ENST00000380249 | + | 9 | 20867003 | 20867212 | 210 | GCCUGAAAAGGUAGGCAUAUCUGCUUUCUCACUGGUAUUUCUUAAUUUGCUAGGGUUUACAACUUUUUCUCUCUCAUCUUAGGAGAUACUUACCUGAUGAAUAUAUACAUAACCCUAGAUCUGUUGUCCAUGCAAGAUACAAAUUCAUACCCAUUAUUUGUUUAUAUGUAAAAGACUGUAGAGGUUUGAAGCAGAUGGAAGACACCUAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
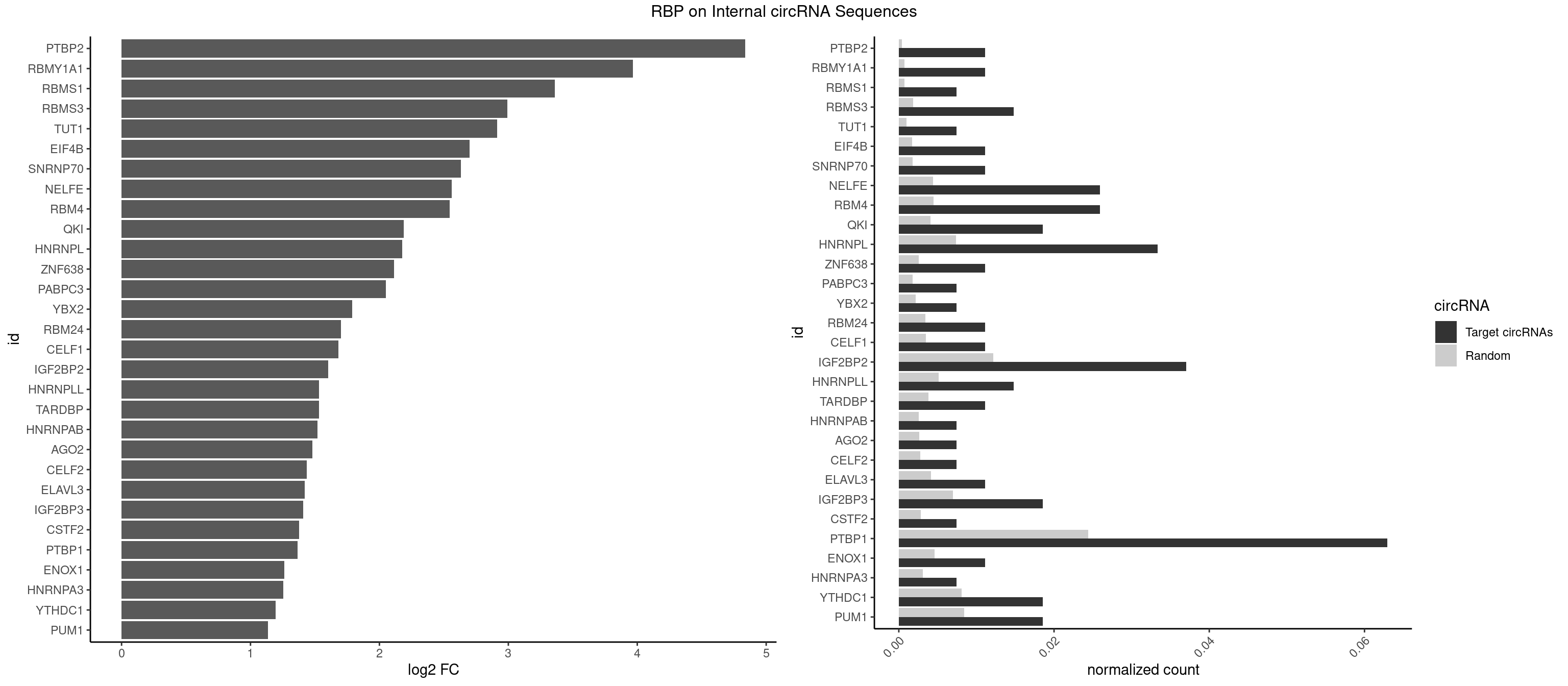
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 2 | 267 | 0.011111111 | 0.0003883867 | 4.838366 | CUCUCU | CUCUCU |
| RBMY1A1 | 2 | 489 | 0.011111111 | 0.0007101099 | 3.967817 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| RBMS1 | 1 | 497 | 0.007407407 | 0.0007217036 | 3.359490 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 | 3 | 1283 | 0.014814815 | 0.0018607779 | 2.993063 | AUAUAG,CAUAUA,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TUT1 | 1 | 678 | 0.007407407 | 0.0009840095 | 2.912225 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| EIF4B | 2 | 1179 | 0.011111111 | 0.0017100607 | 2.699884 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SNRNP70 | 2 | 1237 | 0.011111111 | 0.0017941145 | 2.630659 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| NELFE | 6 | 3028 | 0.025925926 | 0.0043896388 | 2.562221 | CUCUCU,CUCUGG,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 6 | 3065 | 0.025925926 | 0.0044432593 | 2.544705 | CCUUCC,CUUCCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| QKI | 4 | 2805 | 0.018518519 | 0.0040664663 | 2.187121 | ACACAC,ACUCAU,CUCAUA,UACUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPL | 8 | 5085 | 0.033333333 | 0.0073706513 | 2.177102 | AAACAA,AACACA,AAUACA,ACACAA,ACACAC,ACACCA,CACAAG,CACACC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| ZNF638 | 2 | 1773 | 0.011111111 | 0.0025708878 | 2.111665 | GGUUGU,GUUGUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPC3 | 1 | 1234 | 0.007407407 | 0.0017897669 | 2.049197 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| YBX2 | 1 | 1480 | 0.007407407 | 0.0021462711 | 1.787136 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM24 | 2 | 2357 | 0.011111111 | 0.0034172229 | 1.701107 | GAGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 | 2 | 2391 | 0.011111111 | 0.0034664959 | 1.680453 | GUUGUG,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| IGF2BP2 | 9 | 8408 | 0.037037037 | 0.0121863560 | 1.603702 | AAAACA,AACACA,AAUACA,ACACAC,CAAAAC,CAAUAC,CCAUUC,GAACAC,GCAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| HNRNPLL | 3 | 3534 | 0.014814815 | 0.0051229360 | 1.531998 | ACACAC,ACACCA,CACACC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| TARDBP | 2 | 2654 | 0.011111111 | 0.0038476365 | 1.529959 | GUUGUG,GUUGUU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPAB | 1 | 1782 | 0.007407407 | 0.0025839306 | 1.519401 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| AGO2 | 1 | 1830 | 0.007407407 | 0.0026534924 | 1.481076 | UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CELF2 | 1 | 1886 | 0.007407407 | 0.0027346479 | 1.437614 | GUUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ELAVL3 | 2 | 2867 | 0.011111111 | 0.0041563169 | 1.418626 | AUUUUA,UAUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| IGF2BP3 | 4 | 4815 | 0.018518519 | 0.0069793662 | 1.407801 | AAAACA,AACACA,AAUACA,ACACAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| CSTF2 | 1 | 1967 | 0.007407407 | 0.0028520334 | 1.376978 | UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| PTBP1 | 16 | 16854 | 0.062962963 | 0.0244263326 | 1.366066 | AUUUUC,CCUUCC,CUCUCU,CUUCCU,UAUUUU,UCCUCU,UCUCUC,UUCCUU,UUCUCU,UUCUUC,UUUUCU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| ENOX1 | 2 | 3195 | 0.011111111 | 0.0046316558 | 1.262403 | AAGACA,AAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPA3 | 1 | 2140 | 0.007407407 | 0.0031027457 | 1.255423 | CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| YTHDC1 | 4 | 5576 | 0.018518519 | 0.0080822104 | 1.196147 | GAAUGC,UAGUGC,UCAUAC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| PUM1 | 4 | 5823 | 0.018518519 | 0.0084401638 | 1.133626 | AAUUGU,AGAAUU,AUUGUA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
RBP on BSJ (Exon Only)
Plot
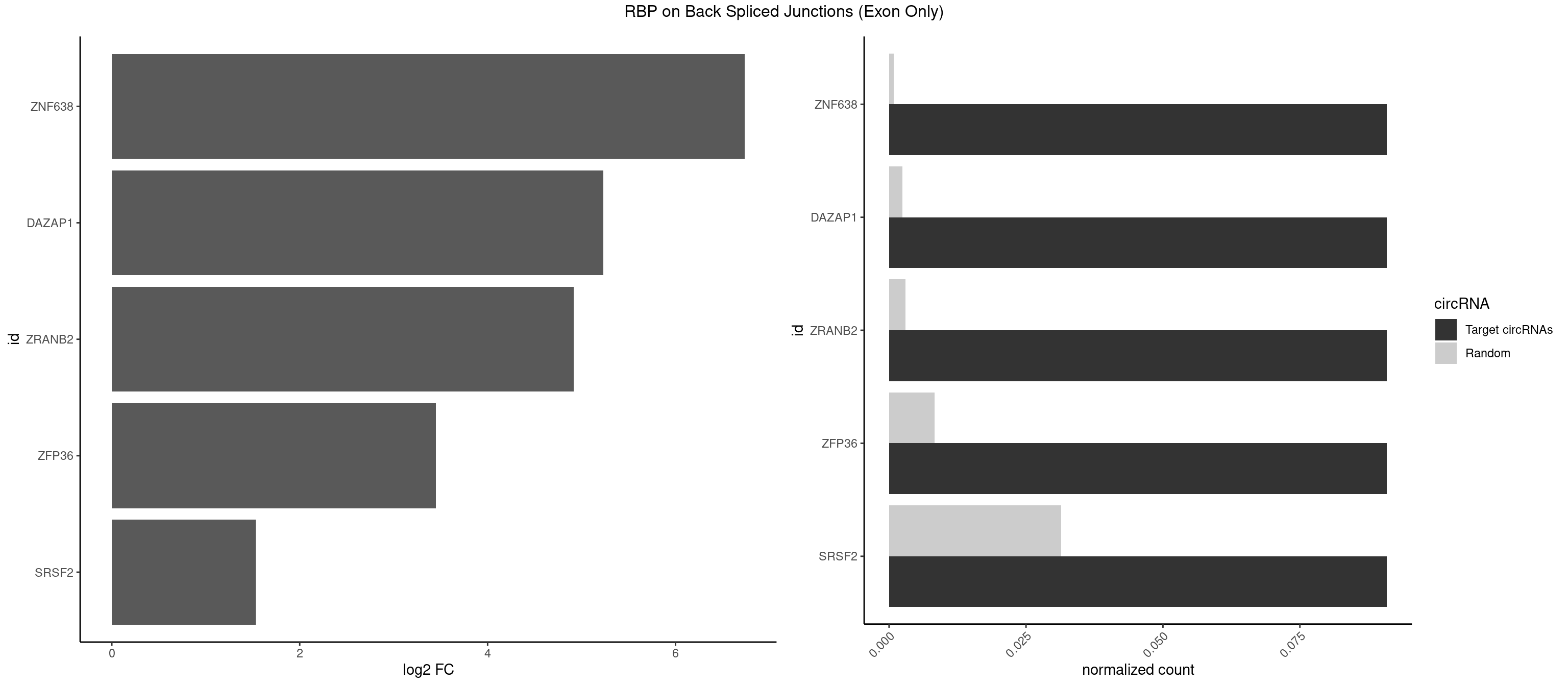
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZNF638 | 1 | 12 | 0.09090909 | 0.000851454 | 6.738352 | GGUUGU | CGUUCG,CGUUCU,CGUUGG,GGUUCU,GGUUGG,GUUCUU,GUUGGU,UGUUCU,UGUUGG,UGUUGU |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | AGGUUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AAAGGU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAAGG | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | GGUUGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
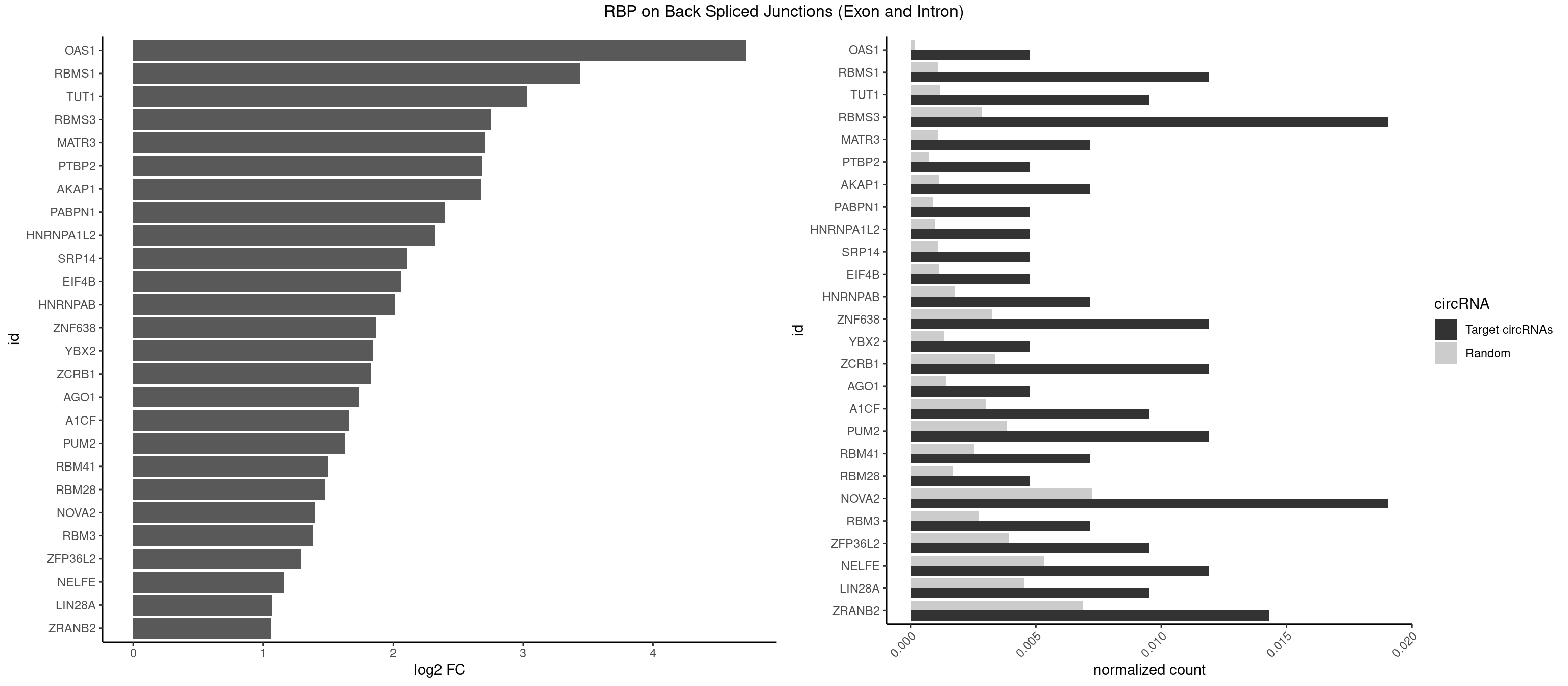
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| RBMS1 | 4 | 217 | 0.011904762 | 0.0010983474 | 3.438132 | AUAUAC,AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| TUT1 | 3 | 230 | 0.009523810 | 0.0011638452 | 3.032640 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBMS3 | 7 | 562 | 0.019047619 | 0.0028365578 | 2.747397 | AAUAUA,AUAUAG,AUAUAU,CAUAUA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAAGAC,AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | GGUUGG,GGUUGU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | AAUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | AGGUAG | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | UACAUA,UAUAUA,UGUAAA,UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACAU,AUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | AGGCAU,CCUAGA,CUAGAU,GAGGCA,UAGAUC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUCU,CUGGUU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | AGGAGA,GGAGAU,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| ZRANB2 | 5 | 1360 | 0.014285714 | 0.0068571141 | 1.058900 | AAAGGU,AGGUAG,AGGUUU,GAGGUU,GGUGGU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.