circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000109674:+:4:177322458:177353728
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000109674:+:4:177322458:177353728 | ENSG00000109674 | ENST00000264596 | + | 4 | 177322459 | 177353728 | 1304 | GCUGCUGCACUGAAUAAUGAUUCCAGCCAGAAUGUCUUGAGCCUGUUUAAUGGAUAUGUUUACAGUGGCGUGGAAACUUUGGGGAAGGAGCUCUUUAUGUACUUUGGACCAAAAGCUUUACGGAUUCAUUUCGGAAUGAAAGGCUUCAUCAUGAUUAAUCCACUUGAGUAUAAAUAUAAAAAUGGAGCUUCUCCUGUUUUGGAAGUGCAGCUCACCAAAGAUUUGAUUUGUUUCUUUGACUCAUCAGUAGAACUCAGAAACUCAAUGGAAAGCCAACAGAGAAUAAGAAUGAUGAAAGAAUUAGAUGUAUGUUCACCUGAAUUUAGUUUCUUGAGAGCAGAAAGUGAAGUUAAAAAACAGAAAGGCCGGAUGCUAGGUGAUGUGCUAAUGGAUCAGAACGUAUUGCCUGGAGUAGGGAACAUCAUCAAAAAUGAAGCUCUCUUUGACAGUGGUCUCCACCCAGCUGUUAAAGUUUGUCAAUUAACAGAUGAACAGAUCCAUCACCUCAUGAAAAUGAUACGUGAUUUCAGCAUUCUCUUUUACAGGUGCCGUAAAGCAGGACUUGCUCUCUCUAAACACUAUAAGGUUUACAAGCGUCCUAAUUGUGGUCAGUGCCACUGCAGAAUAACUGUGUGCCGCUUUGGGGACAAUAACAGAAUGACAUAUUUCUGUCCUCACUGUCAAAAAGAAAAUCCUCAACAUGUUGACAUAUGCAAGCUACCGACUAGAAAUACUAUAAUCAGUUGGACAUCUAGCAGGGUGGAUCAUGUUAUGGACUCCGUGGCUCGGAAGUCGGAAGAGCACUGGACCUGUGUGGUGUGUACUUUAAUCAAUAAGCCCUCUUCUAAGGCAUGUGAUGCUUGCUUGACCUCAAGGCCUAUUGAUUCAGUGCUCAAGAGUGAAGAAAAUUCUACUGUCUUUAGCCACUUAAUGAAGUACCCGUGUAAUACUUUUGGAAAACCUCAUACAGAAGUCAAGAUCAACAGAAAAACUGCAUUUGGAACUACAACUCUUGUCUUGACUGAUUUUAGCAAUAAAUCCAGUACUUUGGAAAGAAAAACAAAGCAAAACCAGAUACUAGAUGAGGAGUUUCAAAACUCUCCUCCUGCUAGUGUUUGUUUGAAUGAUAUACAGCACCCCUCCAAGAAGACAACAAACGAUAUAACUCAACCAUCCAGCAAAGUAAACAUAUCACCUACAAUCAGUUCAGAAUCUAAAUUAUUUAGUCCAGCACAUAAAAAACCGAAAACAGCCCAAUACUCAUCACCAGAGCUUAAAAGCUGCAACCCUGGAUAUUCUAACAGGCUGCUGCACUGAAUAAUGAUUCCAGCCAGAAUGUCUUGAGCCUGUUUAA | circ |
| ENSG00000109674:+:4:177322458:177353728 | ENSG00000109674 | ENST00000264596 | + | 4 | 177322459 | 177353728 | 22 | UAUUCUAACAGGCUGCUGCACU | bsj |
| ENSG00000109674:+:4:177322458:177353728 | ENSG00000109674 | ENST00000264596 | + | 4 | 177322259 | 177322468 | 210 | CUCUCUUUAAAUGACUGCAUAGUGAUUUGACAAAAUGUAUGUAAACAGCCGAUUACAUUACAUGACUUGUUGGAGAUUAUUCUUCUCAUUGGAAUGCACCUGGGCGUGAAGUAAUAUAAACACAUUUAAGAAUUAAAUAAUGCUUUUGCUUAGAGUUUGUGUGUGUGUGUGCUUAUGUGUGUACAUGUAUUUUCCACUAGGCUGCUGCAC | ie_up |
| ENSG00000109674:+:4:177322458:177353728 | ENSG00000109674 | ENST00000264596 | + | 4 | 177353719 | 177353928 | 210 | AUUCUAACAGGUAUGAUGCUUUUAACCUUGGUCUUUAAGAUAAUUGCUUUUUUUUUUUUUUUUUUAAGACGAGGUCUCACUCUGUCACCUAGGCUACAGUGCAGUGGUGUGAUCUCAGCUCACUGCAACCUCUGCCUCCCGCGUUCAAGCGAUUCUCCUGCCUCAGCCUCCCAAGUAGCUGAGAUUACAGGCACAUGCCAUCAGGCCCGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
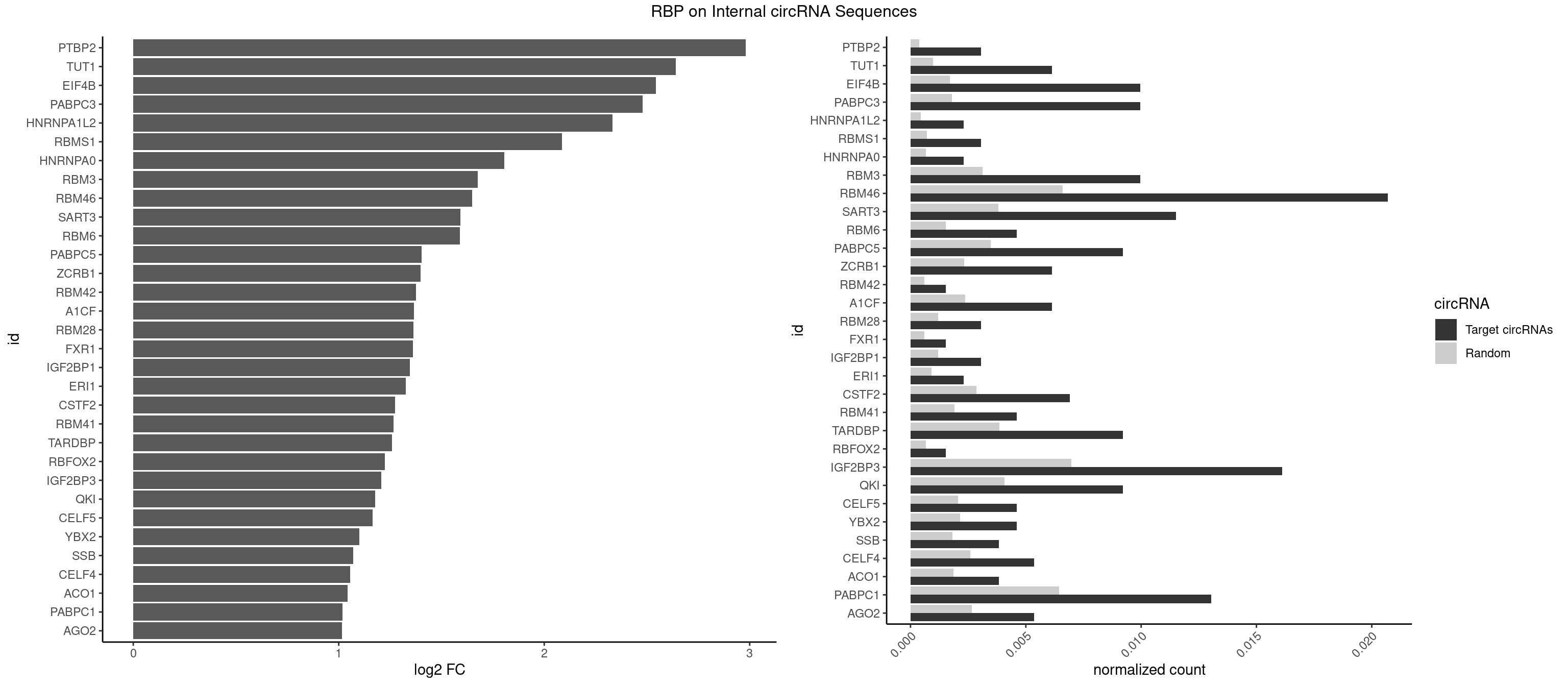
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 3 | 267 | 0.003067485 | 0.0003883867 | 2.981491 | CUCUCU | CUCUCU |
| TUT1 | 7 | 678 | 0.006134969 | 0.0009840095 | 2.640312 | AAAUAC,AAUACU,AGAUAC,CAAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| EIF4B | 12 | 1179 | 0.009969325 | 0.0017100607 | 2.543448 | CUCGGA,GUCGGA,GUUGGA,UCGGAA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC3 | 12 | 1234 | 0.009969325 | 0.0017897669 | 2.477724 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPA1L2 | 2 | 314 | 0.002300613 | 0.0004564992 | 2.333334 | GUAGGG,UAGGGA | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBMS1 | 3 | 497 | 0.003067485 | 0.0007217036 | 2.087578 | AUAUAC,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPA0 | 2 | 453 | 0.002300613 | 0.0006579386 | 1.805994 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBM3 | 12 | 2152 | 0.009969325 | 0.0031201361 | 1.675887 | AAAACU,AAACGA,AAUACU,AUACUA,GAAACU,GAUACG,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RBM46 | 26 | 4554 | 0.020705521 | 0.0066011240 | 1.649232 | AAUCAA,AAUGAA,AAUGAU,AUCAAA,AUCAAU,AUCAUG,AUGAAA,AUGAAG,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SART3 | 14 | 2634 | 0.011503067 | 0.0038186524 | 1.590883 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RBM6 | 5 | 1054 | 0.004601227 | 0.0015289102 | 1.589515 | AAUCCA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PABPC5 | 11 | 2400 | 0.009202454 | 0.0034795387 | 1.403123 | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZCRB1 | 7 | 1605 | 0.006134969 | 0.0023274215 | 1.398324 | AAUUAA,ACUUAA,GAAUUA,GAUUAA,GCUUAA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBM42 | 1 | 407 | 0.001533742 | 0.0005912752 | 1.375154 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| A1CF | 7 | 1642 | 0.006134969 | 0.0023810421 | 1.365463 | AGUAUA,AUCAGU,GAUCAG,UAAUUG,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM28 | 3 | 822 | 0.003067485 | 0.0011926949 | 1.362831 | AGUAGA,AGUAGG,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| FXR1 | 1 | 411 | 0.001533742 | 0.0005970720 | 1.361079 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| IGF2BP1 | 3 | 831 | 0.003067485 | 0.0012057377 | 1.347140 | ACCCGU,AGCACC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ERI1 | 2 | 632 | 0.002300613 | 0.0009173461 | 1.326481 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| CSTF2 | 8 | 1967 | 0.006901840 | 0.0028520334 | 1.274990 | GUGUGU,GUGUUU,GUUUUG,UGUGUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM41 | 5 | 1318 | 0.004601227 | 0.0019115000 | 1.267313 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 11 | 2654 | 0.009202454 | 0.0038476365 | 1.258046 | GAAUGA,GAAUGU,GUGUGU,GUUUUG,UGAAUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBFOX2 | 1 | 452 | 0.001533742 | 0.0006564894 | 1.224213 | UGACUG | UGACUG,UGCAUG |
| IGF2BP3 | 20 | 4815 | 0.016104294 | 0.0069793662 | 1.206278 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUUC,AACUCA,ACAAAC,ACAAUC,CAAUCA,CAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| QKI | 11 | 2805 | 0.009202454 | 0.0040664663 | 1.178243 | ACACUA,ACUCAU,AUCUAA,AUUAAC,CUAACA,CUCAUA,UAAUCA,UACUCA,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CELF5 | 5 | 1415 | 0.004601227 | 0.0020520728 | 1.164937 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 5 | 1480 | 0.004601227 | 0.0021462711 | 1.100186 | AACAUC,ACAACA,ACAACU,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SSB | 4 | 1260 | 0.003834356 | 0.0018274462 | 1.069155 | CUGUUU,GCUGUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CELF4 | 6 | 1782 | 0.005368098 | 0.0025839306 | 1.054844 | GGUGUG,GUGUGG,GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ACO1 | 4 | 1283 | 0.003834356 | 0.0018607779 | 1.043078 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PABPC1 | 16 | 4443 | 0.013036810 | 0.0064402624 | 1.017400 | AAAAAA,AAAAAC,ACAAAC,AGAAAA,CUAACA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| AGO2 | 6 | 1830 | 0.005368098 | 0.0026534924 | 1.016519 | AAAAAA,AAAGUG,AAGUGC,AGUGCU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
RBP on BSJ (Exon Only)
Plot
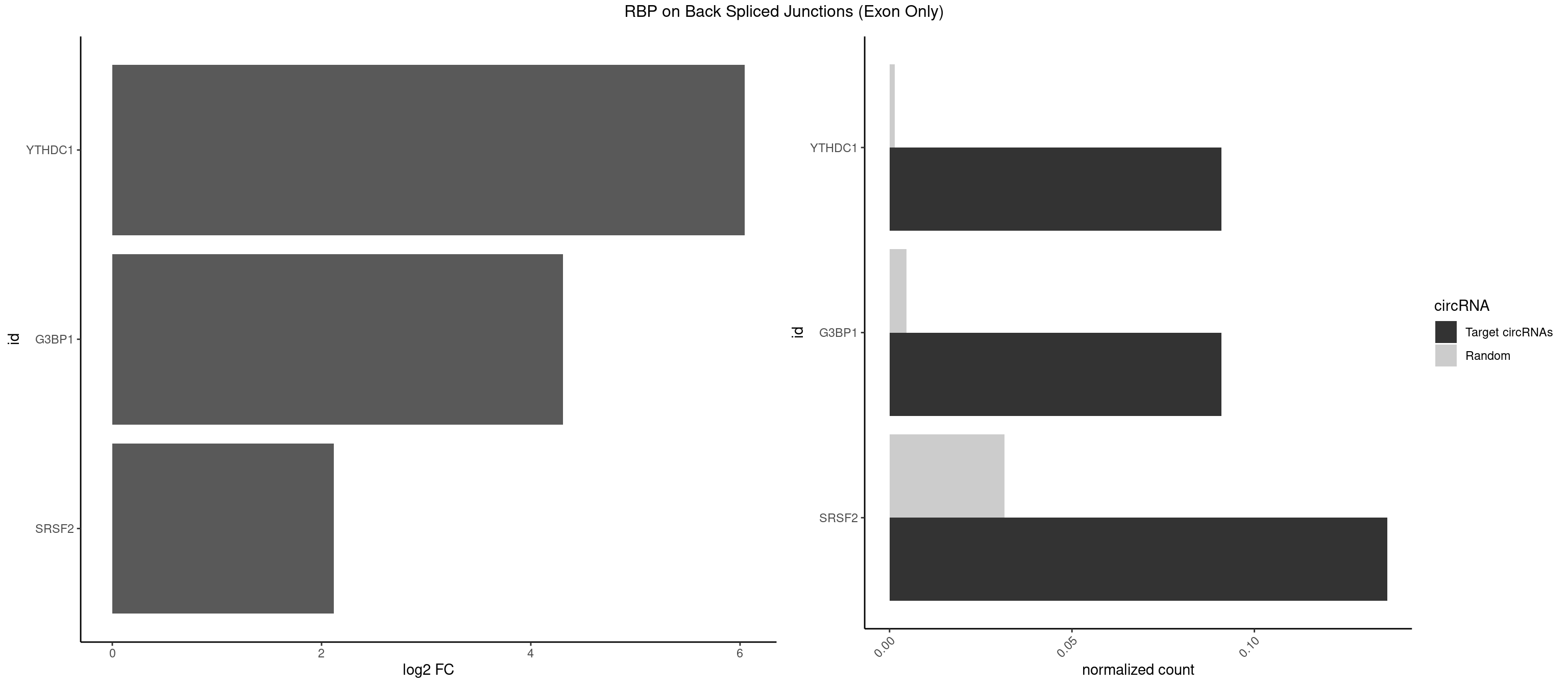
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YTHDC1 | 1 | 20 | 0.09090909 | 0.001375426 | 6.046474 | GGCUGC | GAAUAC,GAGUAC,GCCUGC,GGCUGC,GGGUAC,UAAUAC,UAAUGC,UCAUAC,UCCUGC,UCGUGC,UGAUAC,UGCUGC,UGGUGC |
| G3BP1 | 1 | 69 | 0.09090909 | 0.004584752 | 4.309509 | ACAGGC | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| SRSF2 | 2 | 479 | 0.13636364 | 0.031438302 | 2.116864 | AGGCUG,GGCUGC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
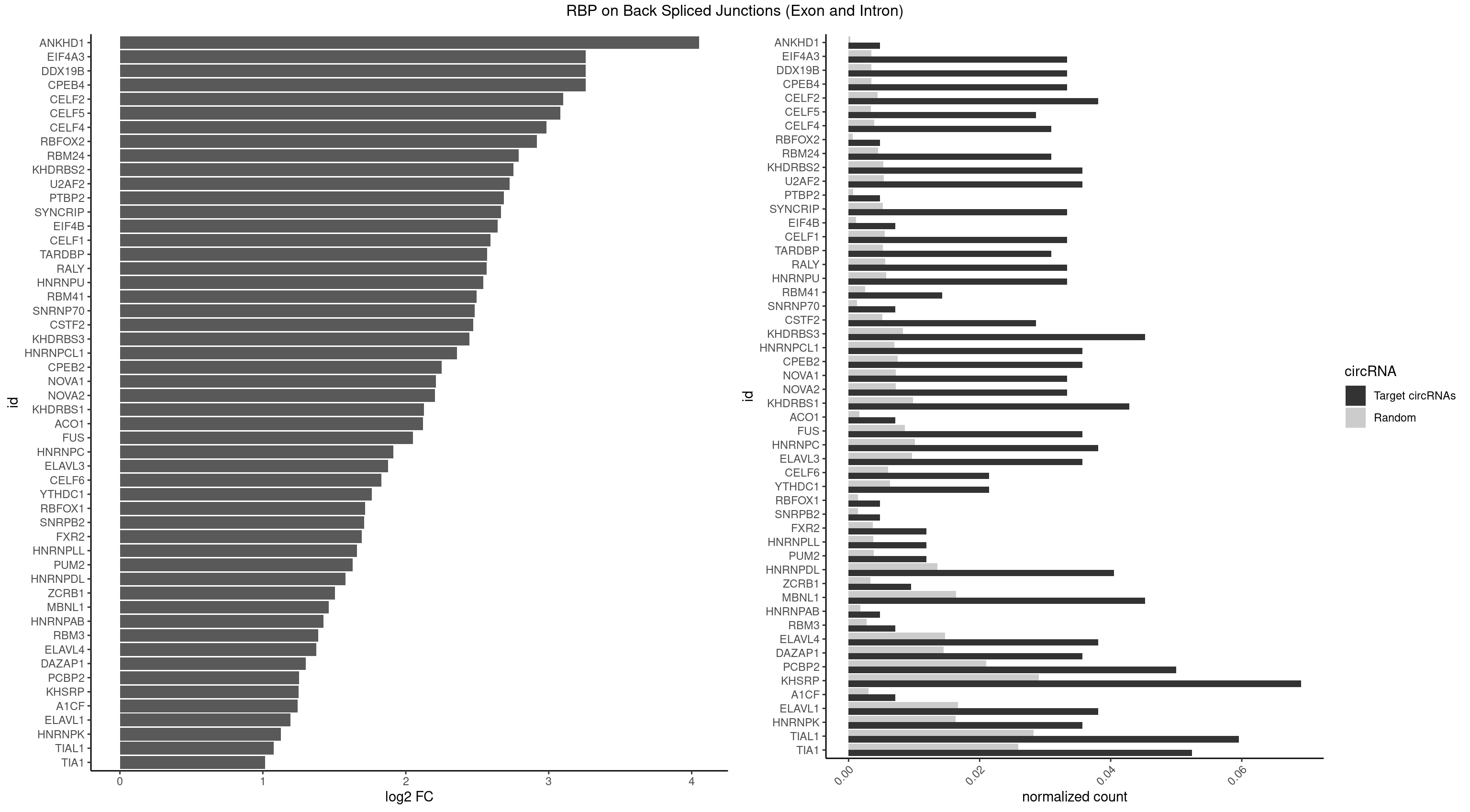
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| CPEB4 | 13 | 691 | 0.033333333 | 0.0034864974 | 3.257115 | UUUUUU | UUUUUU |
| DDX19B | 13 | 691 | 0.033333333 | 0.0034864974 | 3.257115 | UUUUUU | UUUUUU |
| EIF4A3 | 13 | 691 | 0.033333333 | 0.0034864974 | 3.257115 | UUUUUU | UUUUUU |
| CELF2 | 15 | 881 | 0.038095238 | 0.0044437727 | 3.099754 | AUGUGU,GUAUGU,GUGUGU,UAUGUG,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 11 | 669 | 0.028571429 | 0.0033756550 | 3.081334 | GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 12 | 776 | 0.030952381 | 0.0039147521 | 2.983058 | GGUGUG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| RBM24 | 12 | 888 | 0.030952381 | 0.0044790407 | 2.788789 | GUGUGA,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| KHDRBS2 | 14 | 1051 | 0.035714286 | 0.0053002821 | 2.752360 | AUAAAC,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 | 14 | 1071 | 0.035714286 | 0.0054010480 | 2.725190 | UUUUCC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| SYNCRIP | 13 | 1041 | 0.033333333 | 0.0052498992 | 2.666604 | UUUUUU | AAAAAA,UUUUUU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF1 | 13 | 1097 | 0.033333333 | 0.0055320435 | 2.591081 | GUGUGU,GUUUGU,UGUGUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| TARDBP | 12 | 1034 | 0.030952381 | 0.0052146312 | 2.569413 | GUGUGU,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RALY | 13 | 1117 | 0.033333333 | 0.0056328094 | 2.565039 | UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPU | 13 | 1136 | 0.033333333 | 0.0057285369 | 2.540727 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | UACAUG,UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CSTF2 | 11 | 1022 | 0.028571429 | 0.0051541717 | 2.470761 | GUGUGU,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS3 | 18 | 1646 | 0.045238095 | 0.0082980653 | 2.446691 | AAAUAA,AGAUAA,AUAAAC,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPCL1 | 14 | 1381 | 0.035714286 | 0.0069629182 | 2.358737 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 14 | 1487 | 0.035714286 | 0.0074969770 | 2.252120 | CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| NOVA1 | 13 | 1428 | 0.033333333 | 0.0071997179 | 2.210953 | UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 | 13 | 1434 | 0.033333333 | 0.0072299476 | 2.204908 | UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 17 | 1945 | 0.042857143 | 0.0098045143 | 2.128018 | AUUUAA,CAAAAU,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| FUS | 14 | 1711 | 0.035714286 | 0.0086255542 | 2.049812 | UGGUGU,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPC | 15 | 2006 | 0.038095238 | 0.0101118501 | 1.913564 | CUUUUU,UUUUUA,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 14 | 1928 | 0.035714286 | 0.0097188634 | 1.877642 | UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF6 | 8 | 1196 | 0.021428571 | 0.0060308343 | 1.829106 | GUGGUG,UGUGAU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| YTHDC1 | 8 | 1254 | 0.021428571 | 0.0063230552 | 1.760842 | GAAUGC,GACUGC,GGCUAC,GGCUGC,UAAUGC,UCCUGC,UGAUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | AGACGA,GACAAA,GACGAG,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACUGCA,AGACGA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUACAU,UAAAUA,UGUAAA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPDL | 16 | 2689 | 0.040476190 | 0.0135530028 | 1.578461 | AACAGC,AACCUU,ACAUUA,ACCUUG,ACUAGG,ACUGCA,AGUAGC,CACUAG,CUAACA,CUAGGC,CUUUAA,GUAGCU,UAACAG | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,AUUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| MBNL1 | 18 | 3258 | 0.045238095 | 0.0164197904 | 1.462102 | AUGCCA,AUGCUU,CUGCCU,CUGCUG,GCUGCU,GCUUUU,GUGCUU,UGCUGC,UGCUUU,UUGCUU,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ELAVL4 | 15 | 2916 | 0.038095238 | 0.0146966949 | 1.374119 | UAUUUU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| DAZAP1 | 14 | 2878 | 0.035714286 | 0.0145052398 | 1.299927 | AGUUUG,UUUUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| PCBP2 | 20 | 4164 | 0.050000000 | 0.0209844821 | 1.252605 | AAUUAA,ACAUUA,AUUAAA,CCUCCC,CUCCCA,UUAGAG,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| KHSRP | 28 | 5764 | 0.069047619 | 0.0290457477 | 1.249265 | AGCCUC,AUGUAU,AUGUGU,AUUAUU,CAGCCU,CUGCCU,GCCUCC,GUAUGU,GUGUGU,UAUUUU,UGCCUC,UGUAUG,UGUGUA,UGUGUG,UUGUGU | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | UAUUUU,UGAUUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| HNRNPK | 14 | 3244 | 0.035714286 | 0.0163492543 | 1.127276 | UCCCAA,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AUUUUC,CUUUUA,CUUUUG,CUUUUU,UAUUUU,UUAAAU,UUUAAA,UUUUAA,UUUUUA,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUC,CUUUUA,CUUUUG,CUUUUU,UAUUUU,UCUCCU,UUCUCC,UUUUUA,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.