circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000171634:+:17:67975771:67975958
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000171634:+:17:67975771:67975958 | ENSG00000171634 | ENST00000321892 | + | 17 | 67975772 | 67975958 | 187 | ACCUUGCCACCAUGGAAGAAAGAGUACAAAGACGAUAUUAUGAAAAGCUGACGGAAUUUGUGGCAGAUAUGACCAAAAUUUUUGAUAACUGUCGUUACUACAAUCCAAGUGACUCCCCAUUUUACCAGUGUGCAGAAGUUCUCGAAUCAUUCUUUGUACAGAAAUUGAAAGGCUUCAAAGCUAGCAGACCUUGCCACCAUGGAAGAAAGAGUACAAAGACGAUAUUAUGAAAAGCUG | circ |
| ENSG00000171634:+:17:67975771:67975958 | ENSG00000171634 | ENST00000321892 | + | 17 | 67975772 | 67975958 | 22 | AAAGCUAGCAGACCUUGCCACC | bsj |
| ENSG00000171634:+:17:67975771:67975958 | ENSG00000171634 | ENST00000321892 | + | 17 | 67975572 | 67975781 | 210 | AUUUUAGAACCUCAAUCAGAAACAAAAUUGGAAAAGUACUCUUUUGUUCUUUAUGAAUUGCAAAAUAACCUCCAGGGUGAACCAGCCUGGCUCCAGGACUGCUUGCACAGUAAACAUAUAUACUUGUUAGAACUUCGGAGAAUAUUCACAUUGGAAAAACCUUAAAGCUCUGAAAAUGUUUUACAUUUUGUGUUUUUAAGACCUUGCCAC | ie_up |
| ENSG00000171634:+:17:67975771:67975958 | ENSG00000171634 | ENST00000321892 | + | 17 | 67975949 | 67976158 | 210 | AAGCUAGCAGGUGAGCAGAUGGGUUCGGUAUUCUGAAUUAAUUCAACUCUUCACACUCUUUAUACUAUUCUGUCUAACGAUUCAACCAGUGCAGUUUAUGGUAAAUUUAACCUUAAAUAUCUUUAAAAAAAAAAUAAAUAAAUCAAGACUCCAGGGUUAUGAUCUACUGCCAAGGAGAAGUGGACAAAUUGCUGGGUUUGUUGUUGAUCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
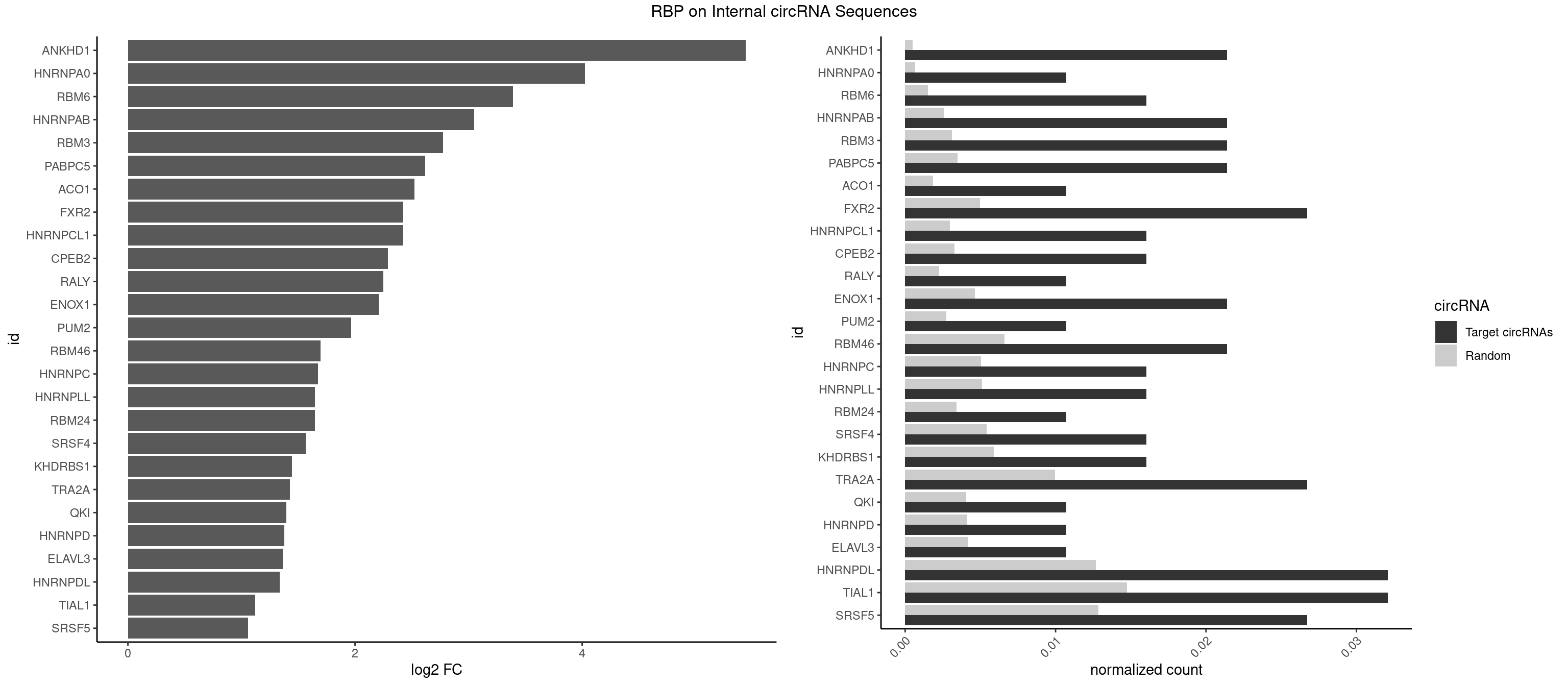
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 3 | 339 | 0.02139037 | 0.0004927293 | 5.440023 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| HNRNPA0 | 1 | 453 | 0.01069519 | 0.0006579386 | 4.022865 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM6 | 2 | 1054 | 0.01604278 | 0.0015289102 | 3.391349 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPAB | 3 | 1782 | 0.02139037 | 0.0025839306 | 3.049322 | AAAGAC,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 3 | 2152 | 0.02139037 | 0.0031201361 | 2.777281 | AAGACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PABPC5 | 3 | 2400 | 0.02139037 | 0.0034795387 | 2.619994 | AGAAAG,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ACO1 | 1 | 1283 | 0.01069519 | 0.0018607779 | 2.522984 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| FXR2 | 4 | 3434 | 0.02673797 | 0.0049780156 | 2.425247 | AGACGA,GACGGA,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPCL1 | 2 | 2062 | 0.01604278 | 0.0029897078 | 2.423848 | AUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 2 | 2261 | 0.01604278 | 0.0032780993 | 2.290993 | AUUUUU,CAUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY | 1 | 1553 | 0.01069519 | 0.0022520629 | 2.247643 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| ENOX1 | 3 | 3195 | 0.02139037 | 0.0046316558 | 2.207362 | AGUACA,GUACAG,UGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM2 | 1 | 1890 | 0.01069519 | 0.0027404447 | 1.964480 | UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM46 | 3 | 4554 | 0.02139037 | 0.0066011240 | 1.696178 | AAUCAU,AUCAUU,AUGAAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPC | 2 | 3471 | 0.01604278 | 0.0050316361 | 1.672825 | AUUUUU,UUUUUG | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPLL | 2 | 3534 | 0.01604278 | 0.0051229360 | 1.646881 | AGACGA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM24 | 1 | 2357 | 0.01069519 | 0.0034172229 | 1.646065 | AGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SRSF4 | 2 | 3740 | 0.01604278 | 0.0054214720 | 1.565168 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| KHDRBS1 | 2 | 4064 | 0.01604278 | 0.0058910141 | 1.445336 | CAAAAU,UUUUAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| TRA2A | 4 | 6871 | 0.02673797 | 0.0099589296 | 1.424827 | AAGAAA,AGAAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| QKI | 1 | 2805 | 0.01069519 | 0.0040664663 | 1.395114 | AAUCAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD | 1 | 2837 | 0.01069519 | 0.0041128408 | 1.378755 | AGAUAU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL3 | 1 | 2867 | 0.01069519 | 0.0041563169 | 1.363584 | AUUUUA | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| HNRNPDL | 5 | 8759 | 0.03208556 | 0.0126950266 | 1.337661 | ACCUUG,AGAUAU,CCUUGC,CUAGCA,CUUGCC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| TIAL1 | 5 | 10182 | 0.03208556 | 0.0147572438 | 1.120501 | AAAUUU,AAUUUU,AUUUUA,AUUUUU,UUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF5 | 4 | 8869 | 0.02673797 | 0.0128544391 | 1.056623 | GAAGAA,GGAAGA,UACAGA,UGCAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
RBP on BSJ (Exon Only)
Plot
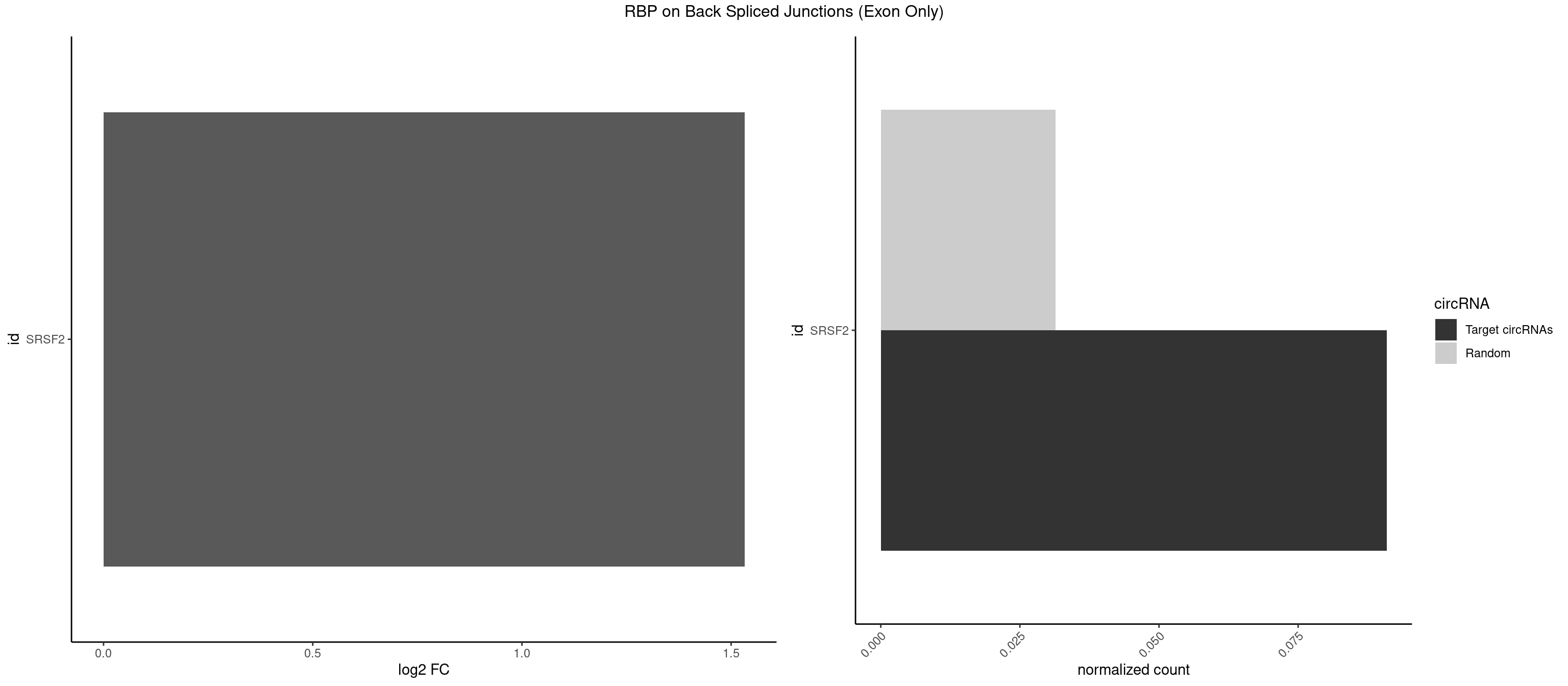
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383 | 1.531901 | AGCAGA | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
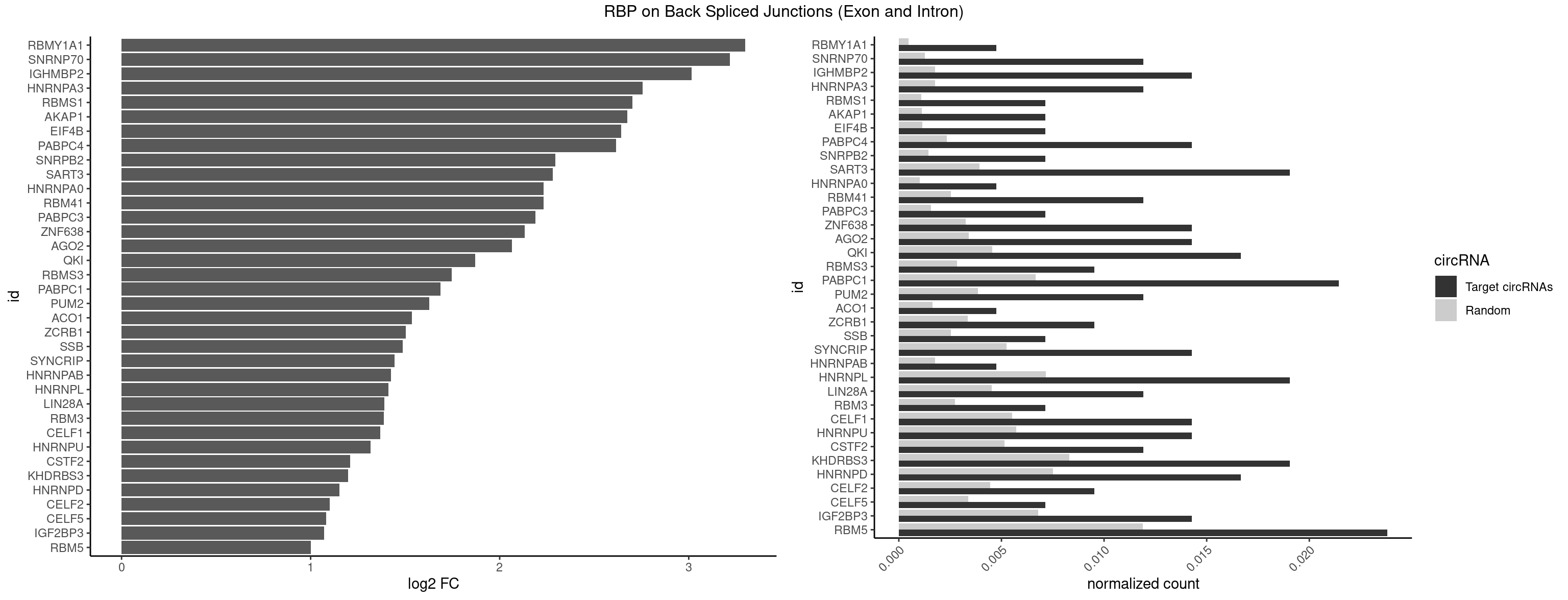
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| SNRNP70 | 4 | 253 | 0.011904762 | 0.0012797259 | 3.217632 | AAUCAA,AUCAAG,AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGHMBP2 | 5 | 350 | 0.014285714 | 0.0017684401 | 3.014024 | AAAAAA | AAAAAA |
| HNRNPA3 | 4 | 349 | 0.011904762 | 0.0017634019 | 2.755106 | AAGGAG,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAC,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC4 | 5 | 462 | 0.014285714 | 0.0023327287 | 2.614483 | AAAAAA | AAAAAA,AAAAAG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | AUUGCA,UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| SART3 | 7 | 777 | 0.019047619 | 0.0039197904 | 2.280762 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACAUU,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAAAC,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZNF638 | 5 | 646 | 0.014285714 | 0.0032597743 | 2.131729 | GGUUCG,GUUCUU,GUUGUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| QKI | 6 | 904 | 0.016666667 | 0.0045596534 | 1.869970 | AUCUAC,CACACU,CUAACG,UAACCU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CAUAUA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| PABPC1 | 8 | 1321 | 0.021428571 | 0.0066606207 | 1.685807 | AAAAAA,AAAAAC,ACAAAU,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAAAU,UAAAUA,UAUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AAUUAA,AUUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | AAAAAA | AAAAAA,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.0071543732 | 1.412713 | AAACAA,AAAUAA,AACAAA,AAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGA,CGGAGA,GGAGAA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACU,AUACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | CUGUCU,GUUUGU,UGUGUU,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUU,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AAAUAA,AAUAAA,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AAAAAA,AAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUUGUU,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,AAAUCA,AAUUCA,ACACUC,CAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | AAAAAA,AAGGAG,AGGGUG,CAAGGA,CUCUUC | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.