circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000183527:-:21:39177434:39181878
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000183527:-:21:39177434:39181878 | ENSG00000183527 | ENST00000331573 | - | 21 | 39177435 | 39181878 | 658 | GGAAGUGCGGCUCCUUCGAAGACAAACAAAAACAUCUUUGGAAGUUUCUUUGCUAGAAAAAUAUCCGUGCUCCAAGUUUAUAAUUGCUAUAGGAAAUAAUGCAGUAGCAUUUCUGUCAUCAUUUGUUAUGAAUUCAGGAGUCUGGGAGGAAGUUGGUUGUGCUAAACUCUGGAAUGAAUGGUGUAGAACAACAGACACUACACAUCUGUCCUCCACAGAGGCUUUUUGUGUGUUUUAUCAUCUAAAAUCCAAUCCCUCGGUUUUUCUCUGUCAGUGCAGUUGCUAUGUUGCAGAAGAUCAACAGUAUCAGUGGCUGGAAAAGGUUUUUGGCUCUUGUCCAAGGAAGAACAUGCAGAUAACUAUUCUCACAUGUCGACAUGUUACCGAUUAUAAAACCUCAGAAUCCACCGGCAGCCUUCCUUCUCCUUUCCUGAGAGCCCUAAAAACACAGAAUUUCAAAGACUCGGCGUGUUGUCCAUUGCUAGAACAACCGAAUAUAGUACACGACCUUCCUGCAGCAGUUCUAAGCUACUGUCAAGUAUGGAAAAUCCCAGCAAUUCUGUACUUGUGUUAUACUGAUGUGAUGAAAUUAGACCUAAUCACAGUGGAAGCUUUUAAGCCUAUACUUUCUACCAGAAGCUUGAAGGGUUUGGUUAAGGGAAGUGCGGCUCCUUCGAAGACAAACAAAAACAUCUUUGGAAGUUUCUU | circ |
| ENSG00000183527:-:21:39177434:39181878 | ENSG00000183527 | ENST00000331573 | - | 21 | 39177435 | 39181878 | 22 | GUUUGGUUAAGGGAAGUGCGGC | bsj |
| ENSG00000183527:-:21:39177434:39181878 | ENSG00000183527 | ENST00000331573 | - | 21 | 39181869 | 39182078 | 210 | GAGUUCUCCGUAAACACGUAUGCAAUAGUUUUAUCUGUUGGUUAUCAUUGUCCGAGUCCAUCGUUUUCUGAGGAUAUUAGUUAUAUUCUUUAUAAAACUAACGUGCAUUUUAUUUAUUGUAAAUCAUACAUAUAGUGCCCUUUUGCCAUGAUACUGUUUAUAUUGAAGUUGCUUUAGUUUCAUCUUGUUUCUUGUGUUAGGGAAGUGCGG | ie_up |
| ENSG00000183527:-:21:39177434:39181878 | ENSG00000183527 | ENST00000331573 | - | 21 | 39177235 | 39177444 | 210 | UUUGGUUAAGGUAAGGUUUUAACUCAUUUAAAUUAAUAGCUGCAUUGUUAUAAUCAGCUUAGUAACAAACAUAAAUGUAUACUGUCAUUUUAAACCCUAAGCAAUGCUUGGAGAUCCAUUCUUCUCAAAGCGAAGCUUUCUAGUCUGGCAGUUUGAAUUUGAUUGUCAUCAUCUCUUCUGUAUACGUCUGUAACUUGACGAAUGCUUUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
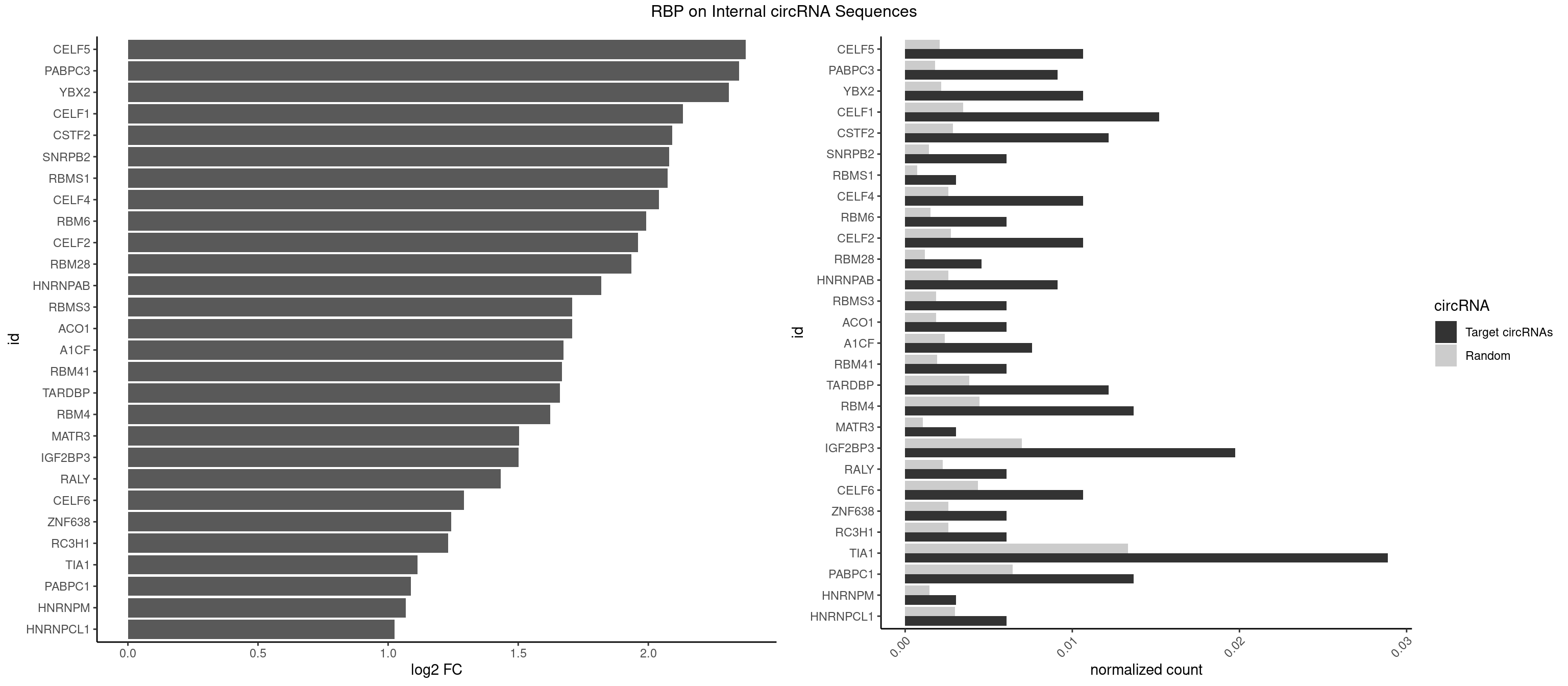
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF5 | 6 | 1415 | 0.010638298 | 0.0020520728 | 2.374114 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC3 | 5 | 1234 | 0.009118541 | 0.0017897669 | 2.349031 | AAAAAC,AAAACA,AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| YBX2 | 6 | 1480 | 0.010638298 | 0.0021462711 | 2.309363 | AACAAC,AACAUC,ACAACA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF1 | 9 | 2391 | 0.015197568 | 0.0034664959 | 2.132291 | GUGUGU,GUGUUG,GUUGUG,UGUGUG,UGUGUU,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| CSTF2 | 7 | 1967 | 0.012158055 | 0.0028520334 | 2.091850 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SNRPB2 | 3 | 991 | 0.006079027 | 0.0014376103 | 2.080168 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 1 | 497 | 0.003039514 | 0.0007217036 | 2.074362 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| CELF4 | 6 | 1782 | 0.010638298 | 0.0025839306 | 2.041628 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM6 | 3 | 1054 | 0.006079027 | 0.0015289102 | 1.991337 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CELF2 | 6 | 1886 | 0.010638298 | 0.0027346479 | 1.959840 | GUGUGU,UAUGUU,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM28 | 2 | 822 | 0.004559271 | 0.0011926949 | 1.934578 | GUGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 5 | 1782 | 0.009118541 | 0.0025839306 | 1.819236 | AAAGAC,AAGACA,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ACO1 | 3 | 1283 | 0.006079027 | 0.0018607779 | 1.707935 | CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 3 | 1283 | 0.006079027 | 0.0018607779 | 1.707935 | AAUAUA,AUAUAG,CUAUAG | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| A1CF | 4 | 1642 | 0.007598784 | 0.0023810421 | 1.674175 | AUAAUU,AUCAGU,CAGUAU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM41 | 3 | 1318 | 0.006079027 | 0.0019115000 | 1.669135 | AUACUU,UACUUG,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TARDBP | 7 | 2654 | 0.012158055 | 0.0038476365 | 1.659868 | GAAUGA,GAAUGG,GUGUGU,GUUGUG,UGAAUG,UGUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM4 | 8 | 3065 | 0.013677812 | 0.0044432593 | 1.622147 | CCUUCC,CCUUCU,CUUCCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| MATR3 | 1 | 739 | 0.003039514 | 0.0010724109 | 1.502983 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IGF2BP3 | 12 | 4815 | 0.019756839 | 0.0069793662 | 1.501184 | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AACACA,AAUUCA,ACAAAC,CAAACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RALY | 3 | 1553 | 0.006079027 | 0.0022520629 | 1.432593 | UUUUUC,UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| CELF6 | 6 | 2997 | 0.010638298 | 0.0043447134 | 1.291934 | GUGAUG,GUGUUG,UGUGAU,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| ZNF638 | 3 | 1773 | 0.006079027 | 0.0025708878 | 1.241574 | GGUUGU,GUUGGU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 3 | 1786 | 0.006079027 | 0.0025897275 | 1.231040 | CCUUCU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| TIA1 | 18 | 9206 | 0.028875380 | 0.0133428208 | 1.113776 | CCUUUC,CUCCUU,CUUUUA,CUUUUU,GUUUUA,GUUUUU,UCCUUU,UCUCCU,UUCUCC,UUUUAU,UUUUCU,UUUUGG,UUUUGU,UUUUUC,UUUUUG | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PABPC1 | 8 | 4443 | 0.013677812 | 0.0064402624 | 1.086646 | AAAAAC,ACAAAC,AGAAAA,CAAACA,CGAAUA,CUAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| HNRNPM | 1 | 999 | 0.003039514 | 0.0014492040 | 1.068580 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| HNRNPCL1 | 3 | 2062 | 0.006079027 | 0.0029897078 | 1.023836 | CUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
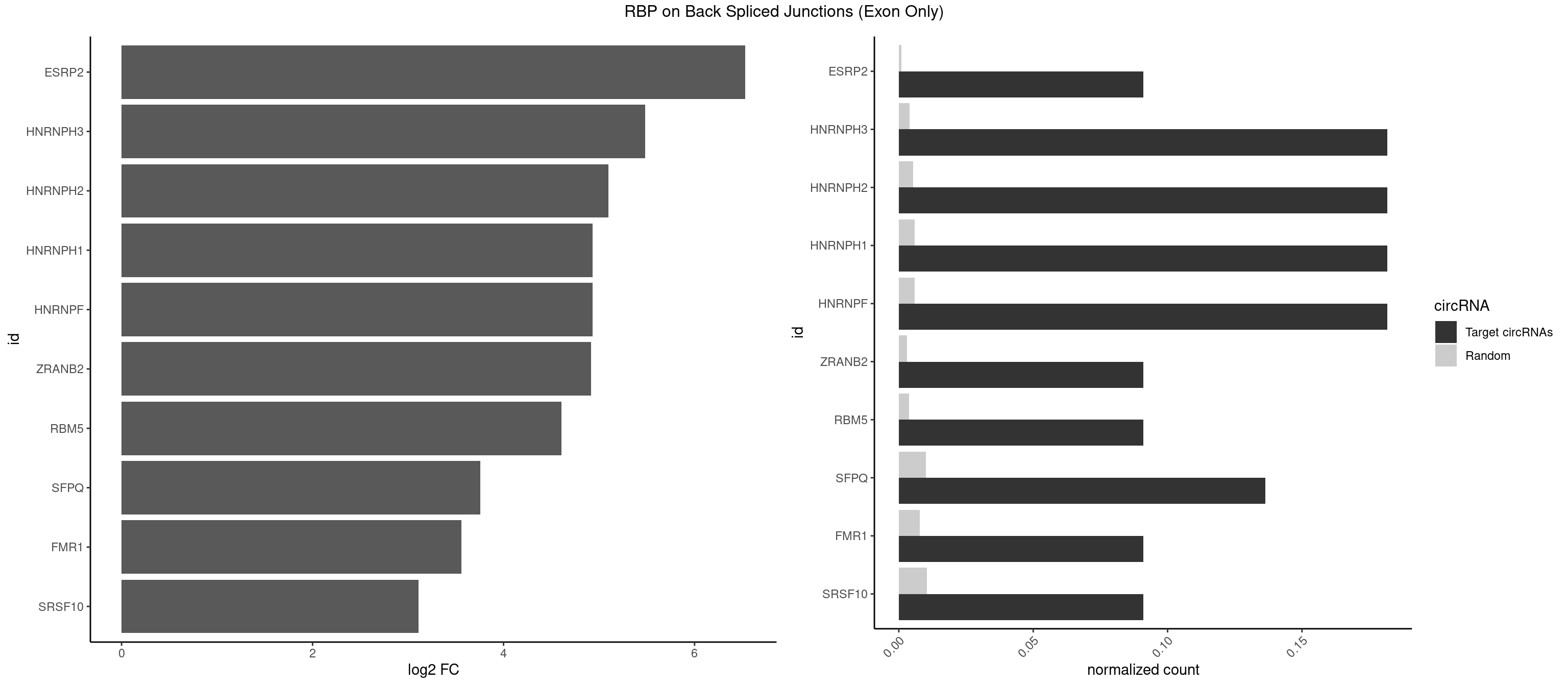
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP2 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | GGGAAG | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,UGGGAA,UGGGGA |
| HNRNPH3 | 3 | 61 | 0.18181818 | 0.0040607807 | 5.484596 | AAGGGA,AGGGAA,GGGAAG | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPH2 | 3 | 80 | 0.18181818 | 0.0053052135 | 5.098942 | AAGGGA,AGGGAA,GGGAAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 3 | 90 | 0.18181818 | 0.0059601782 | 4.930997 | AAGGGA,AGGGAA,GGGAAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 3 | 90 | 0.18181818 | 0.0059601782 | 4.930997 | AAGGGA,AGGGAA,GGGAAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.0030128373 | 4.915230 | AGGGAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| RBM5 | 1 | 56 | 0.09090909 | 0.0037332984 | 4.605902 | AGGGAA | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| SFPQ | 2 | 153 | 0.13636364 | 0.0100864553 | 3.756968 | AAGGGA,UAAGGG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | AAGGGA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | AAGGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
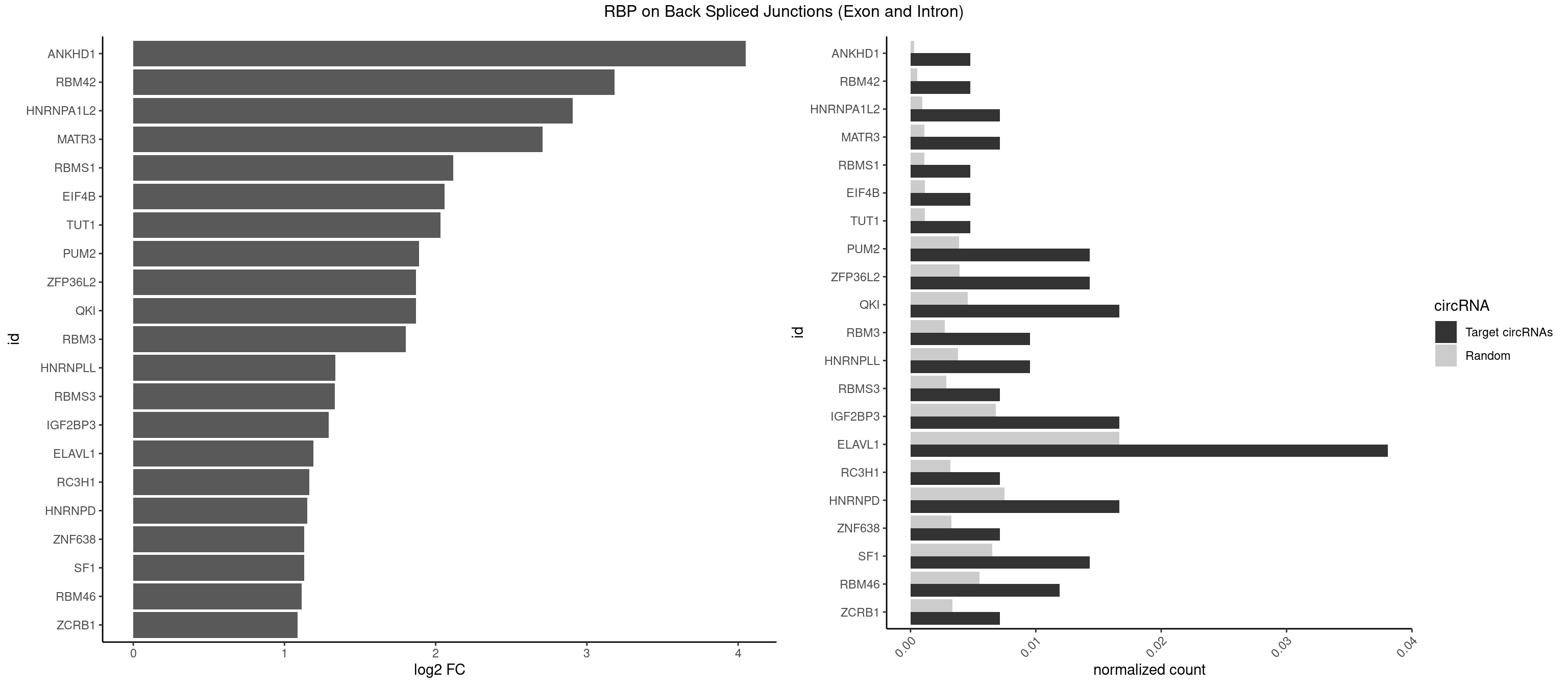
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | GUAAAU,UACAUA,UGUAAA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| QKI | 6 | 904 | 0.016666667 | 0.0045596534 | 1.869970 | AAUCAU,ACUAAC,ACUCAU,AUCAUA,CUAACG,UAAUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AAACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACAAAC,CAAACA,CAUACA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAG,CAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| IGF2BP3 | 6 | 1348 | 0.016666667 | 0.0067966546 | 1.294069 | AAACAC,AAAUCA,AACUCA,ACAAAC,CAAACA,CAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| ELAVL1 | 15 | 3309 | 0.038095238 | 0.0166767432 | 1.191773 | AUUUAU,UAGUUU,UAUUUA,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AUUUAU,UAUUUA,UUAGGG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACUAAC,AGUAAC,UAACAA,UAGUAA,UAUACU | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUCAU,AUCAUA,AUCAUU,AUGAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.