circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000171456:+:20:32366383:32369123
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000171456:+:20:32366383:32369123 | ENSG00000171456 | ENST00000375687 | + | 20 | 32366384 | 32369123 | 195 | GUAUUAGAAAACUACUCGGAUGCUCCAAUGACACCAAAACAGAUUCUGCAGGUCAUAGAGGCAGAAGGACUAAAGGAAAUGAGAAGUGGGACUUCCCCUCUCGCAUGCCUCAAUGCUAUGCUACAUUCCAAUUCAAGAGGAGGAGAGGGGUUGUUUUAUAAACUGCCUGGCCGAAUCAGCCUUUUCACGCUCAAGGUAUUAGAAAACUACUCGGAUGCUCCAAUGACACCAAAACAGAUUCUGCA | circ |
| ENSG00000171456:+:20:32366383:32369123 | ENSG00000171456 | ENST00000375687 | + | 20 | 32366384 | 32369123 | 22 | UCACGCUCAAGGUAUUAGAAAA | bsj |
| ENSG00000171456:+:20:32366383:32369123 | ENSG00000171456 | ENST00000375687 | + | 20 | 32366184 | 32366393 | 210 | AAUUAGAACAUUUCAGUUAAUUGCAGUAACUUUUCAGUAACUAGAUAUAUGUGUAAUUUGAUUUCUGAAGUAAUUUUAACUGUGGAAAAACUAAGUUGUCACCAGCGGUACCUCAUAGCAUAACUUUAGCCCAUGAUAUAUGGAUGGAUGGAUAUAAAUGGAAAUUGCACACUGAAAUUAGGACGUUUAUAUUUCUUCAGGUAUUAGAAA | ie_up |
| ENSG00000171456:+:20:32366383:32369123 | ENSG00000171456 | ENST00000375687 | + | 20 | 32369114 | 32369323 | 210 | CACGCUCAAGGUAAGUGAUAUGAACUCUCUUUUGGUGCAGUGAUUCUUGGACUUUUAAUGUGCAUAAAAAUCACCUGGUAGGUCUGGAAUAGGGGCCAUGAAUUUGCAUUUCUCAUAUGCAGUCAGGUGACACUGAUGUUUUUCUUUUUUUUAGACAUAGUCUCGCUGUGUCACCCAGGCUGGAGUGCAAUGGGGCGAGAUCUCGGCUCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
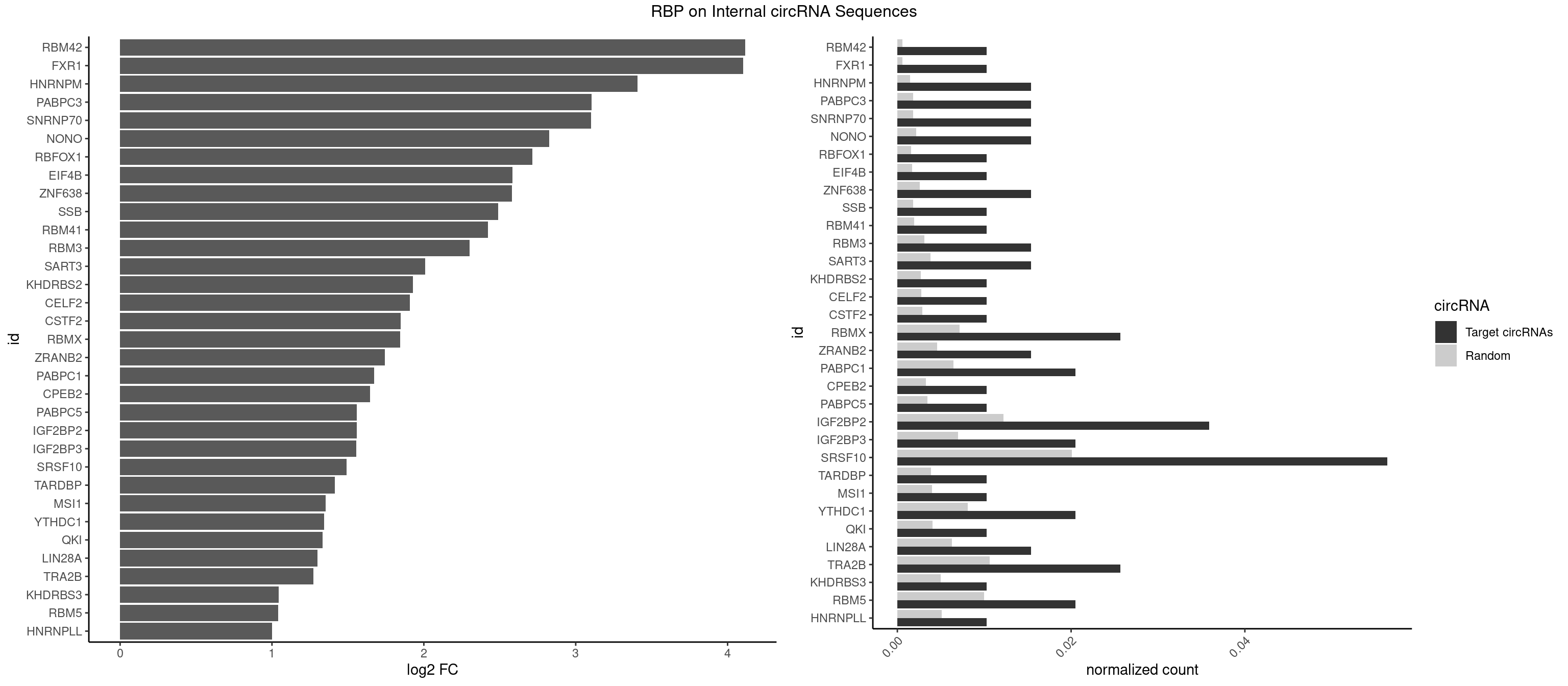
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 407 | 0.01025641 | 0.0005912752 | 4.116552 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.01025641 | 0.0005970720 | 4.102477 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPM | 2 | 999 | 0.01538462 | 0.0014492040 | 3.408156 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| PABPC3 | 2 | 1234 | 0.01538462 | 0.0017897669 | 3.103645 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 2 | 1237 | 0.01538462 | 0.0017941145 | 3.100145 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| NONO | 2 | 1498 | 0.01538462 | 0.0021723567 | 2.824155 | AGAGGA,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBFOX1 | 1 | 1077 | 0.01025641 | 0.0015622419 | 2.714836 | GCAUGC | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| EIF4B | 1 | 1179 | 0.01025641 | 0.0017100607 | 2.584406 | CUCGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZNF638 | 2 | 1773 | 0.01538462 | 0.0025708878 | 2.581150 | GGUUGU,GUUGUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SSB | 1 | 1260 | 0.01025641 | 0.0018274462 | 2.488625 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM41 | 1 | 1318 | 0.01025641 | 0.0019115000 | 2.423749 | UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM3 | 2 | 2152 | 0.01538462 | 0.0031201361 | 2.301808 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SART3 | 2 | 2634 | 0.01538462 | 0.0038186524 | 2.010353 | AGAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| KHDRBS2 | 1 | 1858 | 0.01025641 | 0.0026940701 | 1.928667 | AUAAAC | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CELF2 | 1 | 1886 | 0.01025641 | 0.0027346479 | 1.907099 | GUUGUU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CSTF2 | 1 | 1967 | 0.01025641 | 0.0028520334 | 1.846463 | UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBMX | 4 | 4925 | 0.02564103 | 0.0071387787 | 1.844705 | AAGGAA,ACCAAA,AGAAGG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZRANB2 | 2 | 3173 | 0.01538462 | 0.0045997733 | 1.741854 | AGGUAU,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| PABPC1 | 3 | 4443 | 0.02051282 | 0.0064402624 | 1.671335 | AGAAAA,CGAAUC,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CPEB2 | 1 | 2261 | 0.01025641 | 0.0032780993 | 1.645594 | CCUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PABPC5 | 1 | 2400 | 0.01025641 | 0.0034795387 | 1.559558 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP2 | 6 | 8408 | 0.03589744 | 0.0121863560 | 1.558614 | AAAACA,AAUUCA,ACAUUC,CAAAAC,CAAUUC,GAAAAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| IGF2BP3 | 3 | 4815 | 0.02051282 | 0.0069793662 | 1.555358 | AAAACA,AAUUCA,ACAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SRSF10 | 10 | 13860 | 0.05641026 | 0.0200874160 | 1.489666 | AAAGGA,AAGAGG,AAGGAA,AGAGGA,AGAGGG,GAGAAG,GAGAGG,GAGGAG,GAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| TARDBP | 1 | 2654 | 0.01025641 | 0.0038476365 | 1.414481 | GUUGUU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| MSI1 | 1 | 2770 | 0.01025641 | 0.0040157442 | 1.352787 | AGGAGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| YTHDC1 | 3 | 5576 | 0.02051282 | 0.0080822104 | 1.343704 | GCAUGC,GGAUGC,UGCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| QKI | 1 | 2805 | 0.01025641 | 0.0040664663 | 1.334678 | CUACUC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| LIN28A | 2 | 4315 | 0.01538462 | 0.0062547643 | 1.298461 | AGGAGA,GGAGGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| TRA2B | 4 | 7329 | 0.02564103 | 0.0106226650 | 1.271308 | AAGGAA,AGAAGG,AGGAAA,GAAGGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| KHDRBS3 | 1 | 3429 | 0.01025641 | 0.0049707696 | 1.044985 | AUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBM5 | 3 | 6879 | 0.02051282 | 0.0099705232 | 1.040785 | AAGGAA,AAGGUA,GAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| HNRNPLL | 1 | 3534 | 0.01025641 | 0.0051229360 | 1.001483 | ACACCA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
RBP on BSJ (Exon Only)
Plot
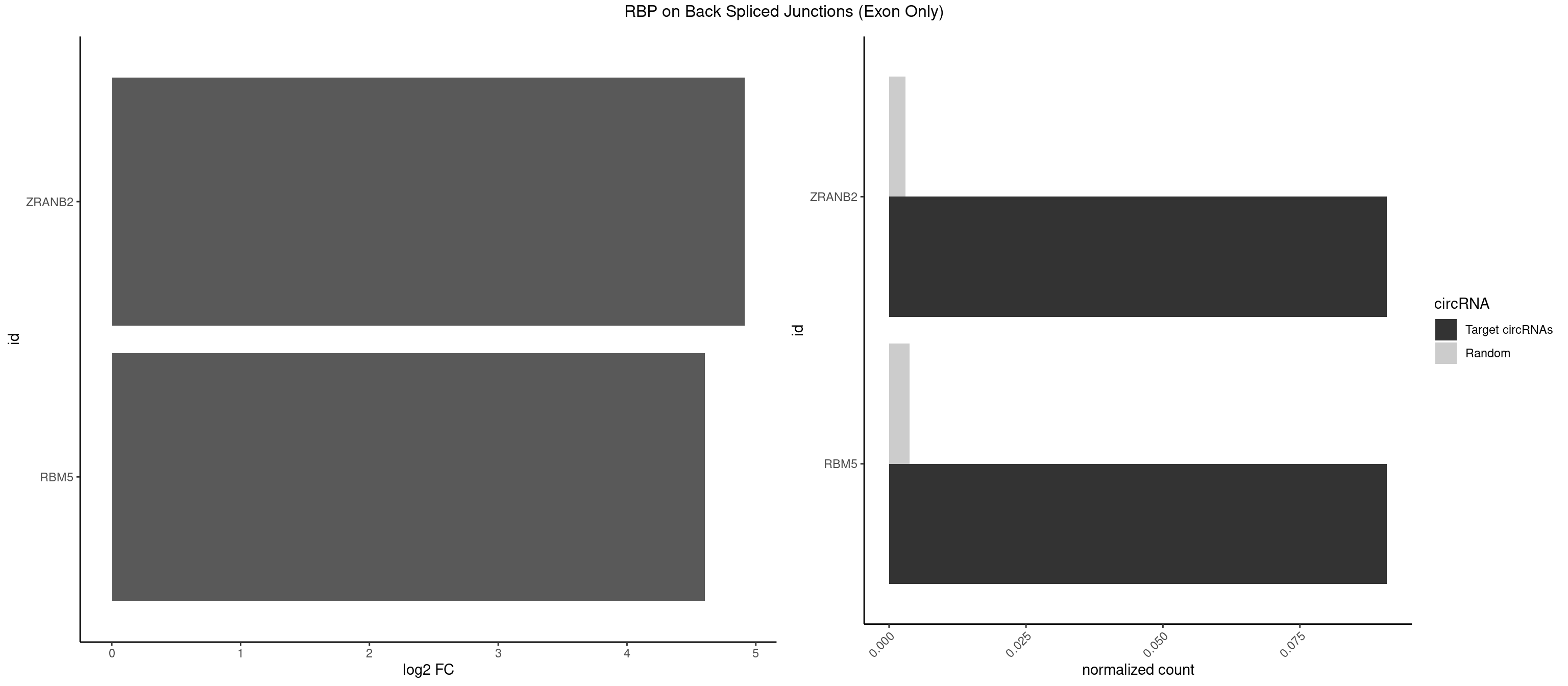
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AGGUAU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | AAGGUA | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
RBP on BSJ (Exon and Intron)
Plot
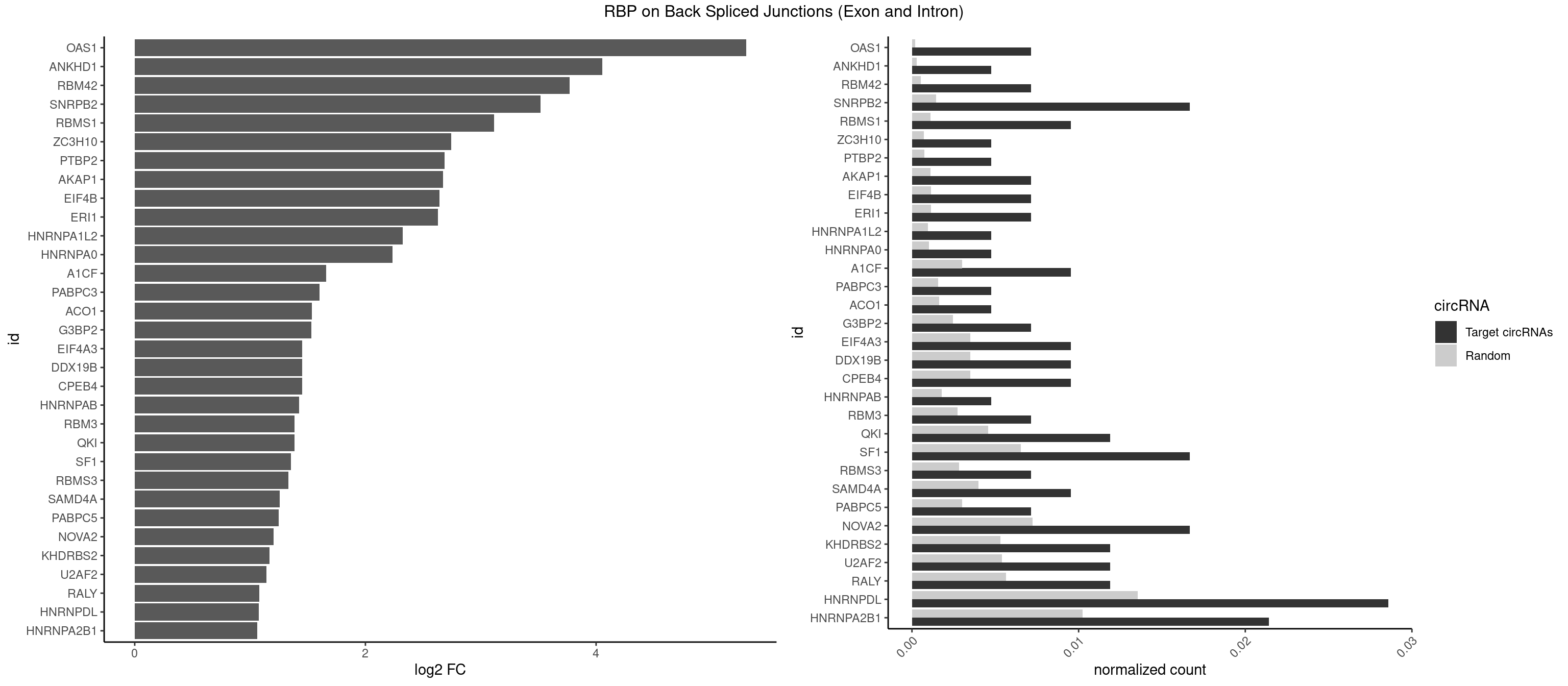
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 2 | 35 | 0.007142857 | 0.0001813785 | 5.299426 | GCAUAA | GCAUAA |
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM42 | 2 | 103 | 0.007142857 | 0.0005239823 | 3.768911 | AACUAA,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| SNRPB2 | 6 | 288 | 0.016666667 | 0.0014560661 | 3.516818 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 3 | 217 | 0.009523810 | 0.0010983474 | 3.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU | AUAUAU,UAUAUA |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| A1CF | 3 | 598 | 0.009523810 | 0.0030179363 | 1.657976 | UAAUUG,UCAGUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | CACACU,CUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACUAAG,AGUAAC,CACUGA,CAGUCA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAA,CUGGUA,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AGACAU,AUCACC,CUAGAU,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPDL | 11 | 2689 | 0.028571429 | 0.0135530028 | 1.075961 | AACUAA,ACUAAG,ACUAGA,ACUUUA,AGAUAU,AUUAGG,CACCAG,CUAAGU,CUAGAU,CUUUAG,UUAGCC | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| HNRNPA2B1 | 8 | 2033 | 0.021428571 | 0.0102478839 | 1.064210 | ACUAGA,AGAUAU,AGGGGC,AUAGCA,AUAGGG,GGGGCC,UAGACA,UUUAUA | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.