circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000112118:-:6:52269085:52279599
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000112118:-:6:52269085:52279599 | ENSG00000112118 | ENST00000616552 | - | 6 | 52269086 | 52279599 | 1437 | GAUGAGGAGAACAAUCCCCUUGAGACAGAAUAUGGCCUUUCUGUCUACAAGGAUCACCAGACCAUCACCAUCCAGGAGAUGCCGGAGAAGGCCCCAGCCGGCCAGCUCCCCCGCUCUGUGGACGUCAUUCUGGAUGAUGACUUGGUGGAUAAAGCGAAGCCUGGUGACCGGGUUCAGGUGGUGGGAACCUACCGUUGCCUUCCUGGAAAGAAGGGAGGCUACACCUCUGGGACCUUCAGGACUGUCCUGAUUGCCUGUAAUGUUAAGCAGAUGAGCAAGGAUGCUCAGCCCUCUUUCUCUGCUGAGGAUAUAGCCAAGAUCAAGAAGUUCAGUAAAACCCGAUCCAAGGAUAUCUUUGACCAGCUGGCCAAGUCAUUGGCCCCAAGUAUCCAUGGGCAUGACUAUGUCAAGAAAGCAAUCCUCUGCUUGCUCUUGGGAGGGGUGGAACGAGACCUAGAAAAUGGCAGCCACAUCCGUGGGGACAUCAAUAUUCUUCUAAUAGGAGACCCAUCCGUUGCCAAGUCUCAGCUUCUGCGGUAUGUGCUUUGCACUGCACCCCGAGCUAUCCCCACCACUGGCCGGGGCUCCUCUGGAGUGGGUCUGACGGCUGCUGUCACCACAGACCAGGAAACAGGAGAGCGCCGUCUGGAAGCAGGGGCCAUGGUCCUGGCUGACCGAGGCGUGGUUUGCAUUGAUGAAUUUGACAAAAUGUCUGACAUGGAUCGCACAGCCAUCCAUGAAGUGAUGGAGCAGGGUCGAGUGACCAUUGCCAAGGCUGGCAUCCAUGCUCGGCUGAAUGCCCGCUGCAGUGUUUUGGCAGCUGCCAACCCUGUCUACGGCAGGUAUGACCAGUAUAAGACUCCAAUGGAGAACAUUGGGCUACAGGACUCACUGCUGUCACGAUUUGACUUGCUCUUCAUCAUGCUGGAUCAGAUGGAUCCUGAGCAGGAUCGGGAGAUCUCAGACCAUGUCCUUCGGAUGCACCGUUACAGAGCACCUGGGGAGCAGGAUGGCGAUGCUAUGCCCUUGGGUAGUGCUGUGGAUAUCCUGGCCACAGAUGAUCCCAACUUUAGCCAGGAAGAUCAGCAGGACACCCAGAUUUAUGAGAAGCAUGACAACCUUCUACAUGGGACCAAGAAGAAAAAGGAGAAGAUGGUGAGUGCAGCAUUCAUGAAGAAGUACAUCCAUGUGGCCAAAAUCAUCAAGCCUGUCCUGACACAGGAGUCGGCCACCUACAUUGCAGAAGAGUAUUCACGCCUGCGCAGCCAGGAUAGCAUGAGCUCAGACACCGCCAGGACAUCUCCAGUUACAGCCCGAACACUGGAAACUCUGAUUCGACUGGCCACAGCCCAUGCGAAGGCCCGCAUGAGCAAGACUGUGGACCUGCAGGAUGCAGAGGAAGCUGUGGAGUUGGUCCAGUAUGCUUACUUUAAGAAGGAUGAGGAGAACAAUCCCCUUGAGACAGAAUAUGGCCUUUCUGUCUACAA | circ |
| ENSG00000112118:-:6:52269085:52279599 | ENSG00000112118 | ENST00000616552 | - | 6 | 52269086 | 52279599 | 22 | ACUUUAAGAAGGAUGAGGAGAA | bsj |
| ENSG00000112118:-:6:52269085:52279599 | ENSG00000112118 | ENST00000616552 | - | 6 | 52279590 | 52279799 | 210 | UUCAGACUAUGGGUUGCCCCUUCCAUUCGAUGGUUGUUACUAAAACAGACAGGAACACAAAAAGCUAGAGAAGGUACUUGGCUUUUUUUGUUACUGAAUAAUAAAAUUUUAUGAAGUCAUCUUAUUUGCCAUGACAGGUGAGUGUGGCAUUUUUAAGACAGGACGGAGAUCACUCUGUCCCUCUGUGUGCCUUUUUCUAGGAUGAGGAGA | ie_up |
| ENSG00000112118:-:6:52269085:52279599 | ENSG00000112118 | ENST00000616552 | - | 6 | 52268886 | 52269095 | 210 | CUUUAAGAAGGUGAGGAUUCAGAUGCCUGGGCAUGAGAGGAUCUCCUGGAUAUAUUACAGAUGCAUGGGCUUCCCAGCCUUCCGUGGCUGACGGCUACAUUUGGAUUGUCAACAGAAAGCAGUAUUUGUCUUCCAGUCAUCAGUCUGCCUCACUCUUUCCUCUGAAACACUCAGUGAUUCUCAACCCUGACUGCACCAGAGAAUCAGAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
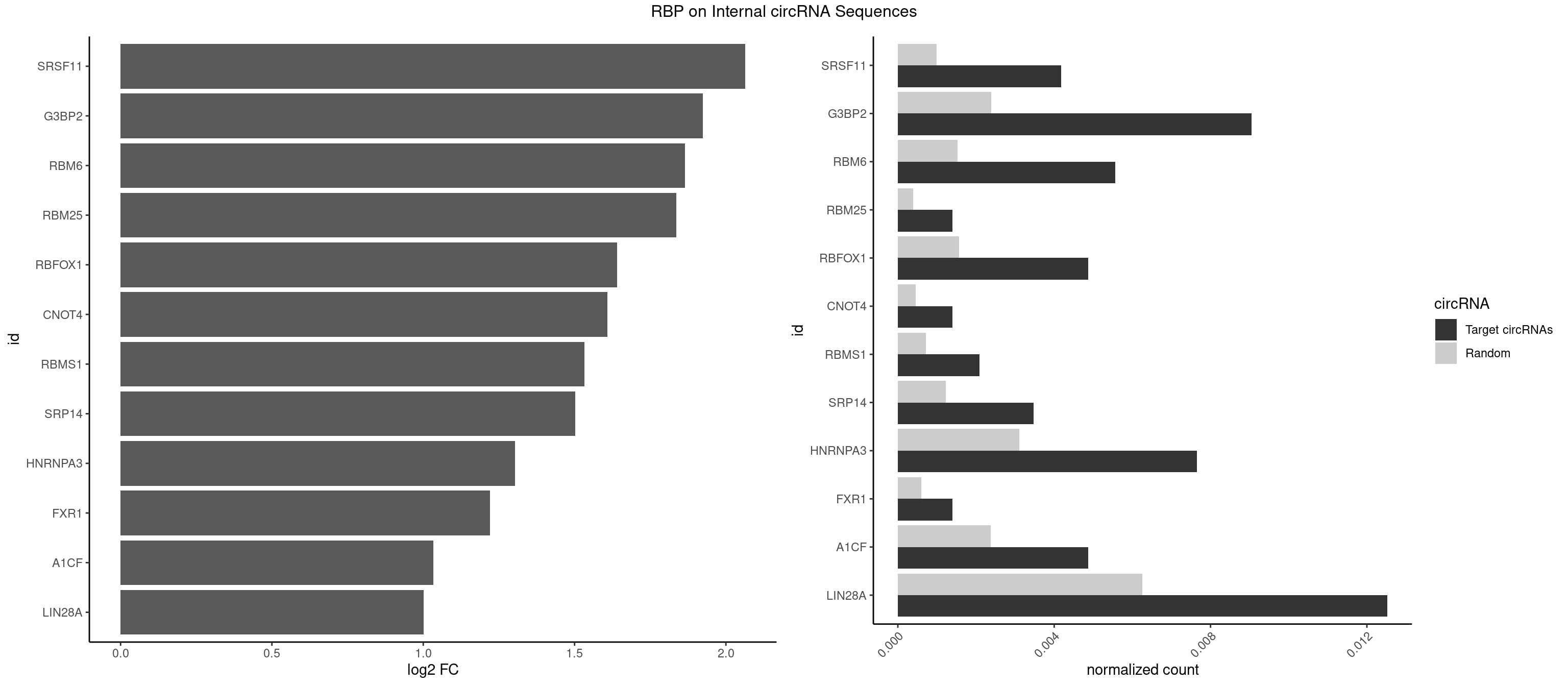
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF11 | 5 | 688 | 0.004175365 | 0.0009985015 | 2.064066 | AAGAAG | AAGAAG |
| G3BP2 | 12 | 1644 | 0.009046625 | 0.0023839405 | 1.924031 | AGGAUA,AGGAUG,GGAUAA,GGAUAG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM6 | 7 | 1054 | 0.005567154 | 0.0015289102 | 1.864436 | AUCCAA,AUCCAG,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM25 | 1 | 268 | 0.001391788 | 0.0003898359 | 1.836001 | AUCGGG | AUCGGG,CGGGCA,UCGGGC |
| RBFOX1 | 6 | 1077 | 0.004871260 | 0.0015622419 | 1.640677 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| CNOT4 | 1 | 314 | 0.001391788 | 0.0004564992 | 1.608256 | GACAGA | GACAGA |
| RBMS1 | 2 | 497 | 0.002087683 | 0.0007217036 | 1.532424 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 4 | 847 | 0.003479471 | 0.0012289250 | 1.501471 | CCUGUA,CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPA3 | 10 | 2140 | 0.007654836 | 0.0031027457 | 1.302826 | AAGGAG,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| FXR1 | 1 | 411 | 0.001391788 | 0.0005970720 | 1.220963 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| A1CF | 6 | 1642 | 0.004871260 | 0.0023810421 | 1.032702 | AGUAUA,CAGUAU,GAUCAG,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| LIN28A | 17 | 4315 | 0.012526096 | 0.0062547643 | 1.001909 | AGGAGA,AGGAGU,CGGAGA,GGAGAA,GGAGAU,GGAGGG,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
RBP on BSJ (Exon Only)
Plot
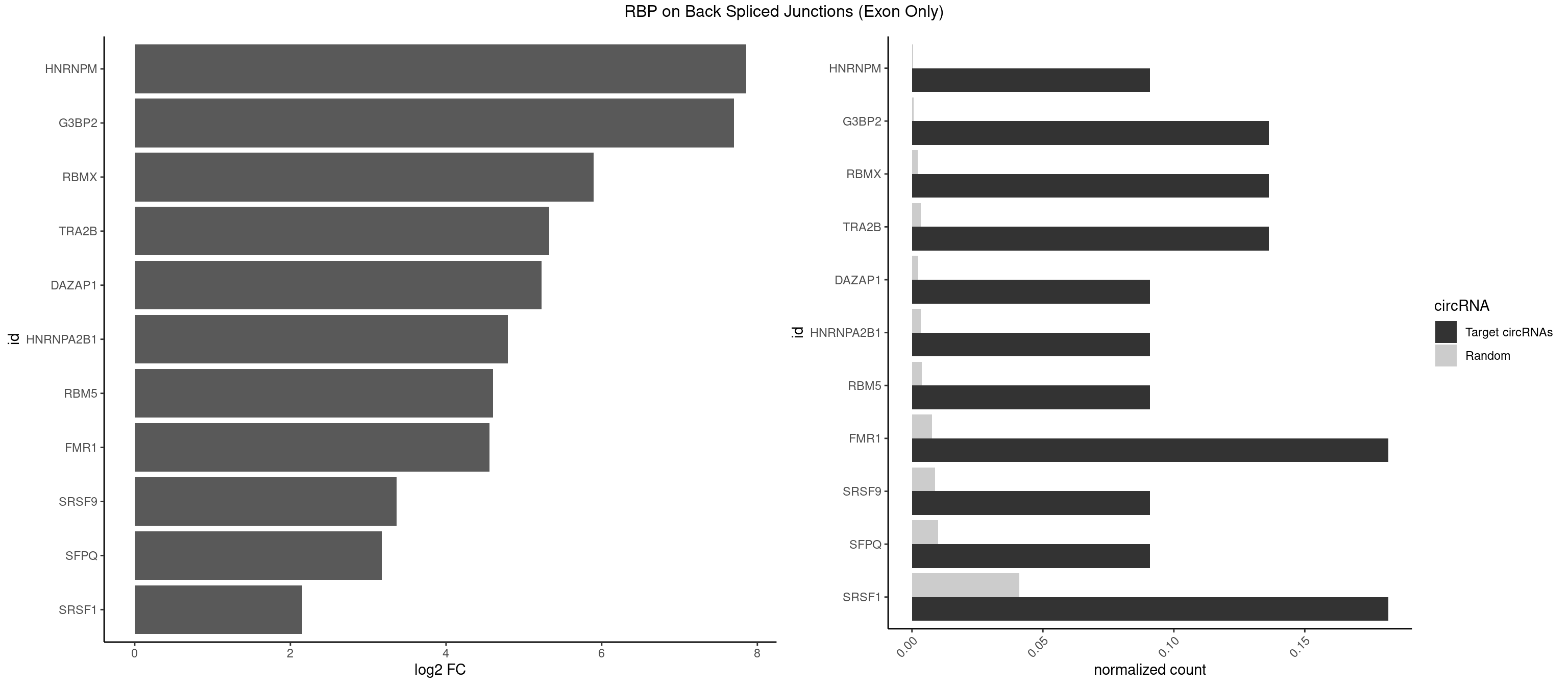
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPM | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GAAGGA | AAGGAA,GAAGGA |
| G3BP2 | 2 | 9 | 0.13636364 | 0.0006549646 | 7.701826 | AGGAUG,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUGA,GGAUGG,GGAUUG |
| RBMX | 2 | 34 | 0.13636364 | 0.0022923762 | 5.894471 | AGAAGG,GAAGGA | AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUGUU,AUCAAA,AUCCCA,GAAGGA,GGAAGG,UAAGAC,UCAAAA |
| TRA2B | 2 | 51 | 0.13636364 | 0.0034058161 | 5.323315 | AGAAGG,GAAGGA | AAAGAA,AAGAAC,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,GAAAGA,GAAGAA,GAAGGA,GGAAGG,UAAGAA |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGGAUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | GAAGGA | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| RBM5 | 1 | 56 | 0.09090909 | 0.0037332984 | 4.605902 | GAAGGA | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| FMR1 | 3 | 117 | 0.18181818 | 0.0077285827 | 4.556149 | AAGGAU,AGGAUG,GAAGGA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF9 | 1 | 134 | 0.09090909 | 0.0088420225 | 3.361976 | GAAGGA | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | GAAGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF1 | 3 | 625 | 0.18181818 | 0.0410007860 | 2.148773 | AGAAGG,GAAGGA,GGAUGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
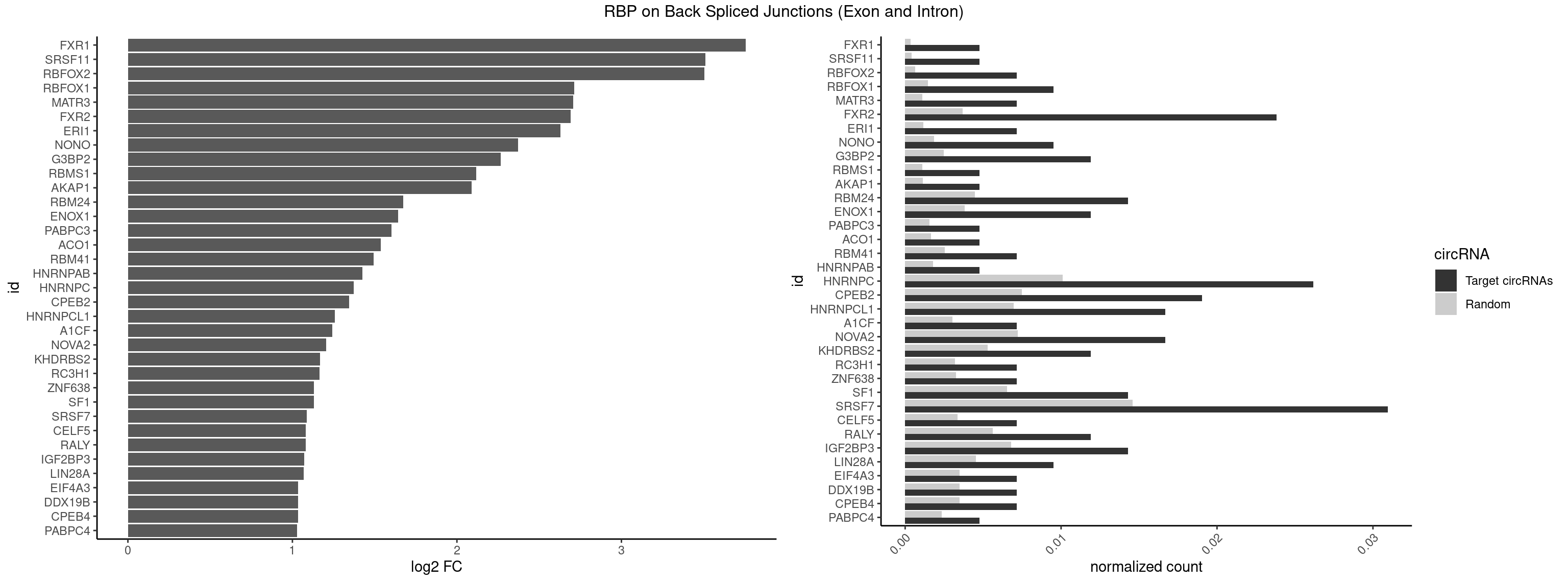
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGACUG,UGCAUG | UGACUG,UGCAUG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | GCAUGA,UGACUG,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| FXR2 | 9 | 730 | 0.023809524 | 0.0036829907 | 2.692589 | AGACAG,GACAGG,GACGGA,GGACGG,UGACAG,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA | UUCAGA,UUUCAG |
| NONO | 3 | 364 | 0.009523810 | 0.0018389762 | 2.372636 | AGAGGA,AGGAAC,GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUG,AGGAUU,GGAUGA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| RBM24 | 5 | 888 | 0.014285714 | 0.0044790407 | 1.673311 | AGUGUG,GAGUGU,GUGUGG,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ENOX1 | 4 | 756 | 0.011904762 | 0.0038139863 | 1.642167 | AAGACA,AGACAG,CAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUU,UACUUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPC | 10 | 2006 | 0.026190476 | 0.0101118501 | 1.372995 | AUUUUU,CUUUUU,GGAUAU,GGAUUC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 7 | 1487 | 0.019047619 | 0.0074969770 | 1.345230 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUCAGU,CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AGAUCA,AGUCAU,GAUCAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCCUUC,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGU,GUUGUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACAGAC,CAGUCA,GCUGAC,UACUAA,UACUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAGAAG,AGACUA,AGAGAA,AGAGGA,AGAUCA,CAGAGA,CUAGAG,GAGAUC,UAGAGA,UCAACA,UGAGAG | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAACA,AAACAC,AACACA,ACACUC,CACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| LIN28A | 3 | 900 | 0.009523810 | 0.0045395002 | 1.069005 | AGGAGA,CGGAGA,GGAGAU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.