circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000100139:+:22:37912350:37932888
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000100139:+:22:37912350:37932888 | ENSG00000100139 | MSTRG.22088.4 | + | 22 | 37912351 | 37932888 | 2039 | GCCUUUGAAGUGGCUGAGAAGGAGCUGGGGAUCCCCGCUCUCCUGGACCCCAAUGACAUGGUCUCCAUGAGCGUCCCUGACUGCCUCAGCAUCAUGACCUAUGUGUCCCAGUAUUACAACCACUUCUGCAGUCCUGGCCAAGCUGGUGUCUCGCCACCCAGAAAGGGCCUUGCACCCUGUUCCCCGCCGUCUGUAGCACCCACUCCAGUGGAACCAGAAGAUGUGGCUCAGGGCGAGGAGCUCUCCUCAGGCAGCCUGUCAGAGCAGGGCACCGGCCAGACCCCCAGCAGCACGUGCGCAGCCUGCCAGCAGCAUGUGCACUUGGUGCAGCGCUACCUGGCUGACGGCAGGCUGUACCAUCGCCACUGCUUCCGGUGUCGGCGGUGCUCCAGCACCCUGCUCCCUGGGGCUUAUGAGAAUGGGCCUGAGGAGGGCACCUUUGUGUGUGCAGAACACUGUGCCAGGCUGGGCCCGGGGACACGGUCGGGGACCAGGCCUGGGCCCUUCUCACAGCCAAAGCAGCAGCACCAGCAGCAACUCGCAGAAGAUGCCAAGGAUGUUCCAGGAGGCGGCCCCAGCUCCAGUGCUCCUGCAGGGGCUGAGGCCGAUGGACCCAAGGCCAGCCCUGAGGCCCGGCCGCAGAUCCCUACCAAGCCCCGGGUUCCUGGCAAACUACAGGAGCUGGCCAGCCCCCCUGCGGGCCGCCCCACCCCUGCCCCCAGGAAGGCCUCUGAGAGCACCACCCCAGCACCCCCCACGCCCCGGCCCCGCUCCAGUCUGCAGCAGGAGAACCUGGUGGAGCAGGCUGGCAGCAGCAGCCUGGUGAACGGGAGACUGCACGAACUGCCUGUCCCCAAGCCGAGGGGGACACCGAAGCCGUCCGAGGGGACACCAGCCCCCAGGAAGGACCCCCCAUGGAUCACGCUGGUGCAGGCAGAACCAAAGAAGAAGCCAGCCCCACUUCCCCCAAGCAGCAGCCCGGGGCCACCAAGCCAGGACAGCAGGCAGGUGGAGAAUGGAGGCACCGAGGAGGUGGCCCAGCCGAGCCCAACGGCCAGCCUGGAGUCCAAACCCUAUAACCCCUUUGAGGAGGAGGAGGAGGACAAGGAGGAAGAGGCUCCAGCUGCACCCAGCCUGGCCACCAGCCCUGCCCUGGGCCACCCGGAGUCCACACCCAAGUCCCUGCACCCCUGGUACGGCAUCACCCCUACCAGCAGCCCCAAGACAAAGAAGCGCCCUGCCCCGCGCGCACCCAGCGCGUCCCCACUGGCUCUCCACGCCUCCCGCCUCUCGCACUCGGAGCCGCCCUCGGCCACACCAUCGCCAGCGCUCAGCGUGGAGAGCCUGUCGUCUGAGAGCGCCAGCCAGACUGCAGGUGCAGAGCUUCUGGAGCCGCCAGCUGUGCCCAAGAGCUCCUCAGAGCCUGCUGUCCAUGCCCCUGGUACCCCUGGAAACCCUGUCAGCCUCUCUACCAACUCCUCCCUGGCCUCCUCUGGGGAACUAGUGGAGCCUAGAGUGGAACAAAUGCCUCAAGCCAGCCCUGGCCUUGCCCCCAGGACCAGGGGCAGCUCAGGUCCCCAGCCAGCCAAGCCCUGCAGUGGCGCCACCCCAACGCCUCUCUUGUUGGUUGGAGACAGGAGCCCGGUGCCUUCCCCUGGAAGCUCGUCCCCACAGCUGCAGGUAAAGUCCUCCUGCAAGGAGAAUCCUUUUAACCGGAAGCCAUCACCUGCAGCGUCCCCAGCCACAAAGAAGGCCACCAAGGGAUCCAAGCCAGUGAGGCCACCUGCCCCUGGACACGGCUUUCCACUCAUCAAACGCAAGGUCCAGGCUGACCAGUACAUCCCUGAGGAGGACAUCCAUGGAGAGAUGGAUACCAUUGAGCGCCGGCUGGAUGCCCUGGAGCACCGUGGGGUGCUGCUGGAGGAGAAGCUGCGUGGCGGCCUGAAUGAGGGCCGUGAGGAUGACAUGCUGGUGGACUGGUUCAAGCUCAUCCACGAGAAGCACCUACUGGUGCGGCGAGAGUCCGAGCUCAUCUAUGUGCCUUUGAAGUGGCUGAGAAGGAGCUGGGGAUCCCCGCUCUCCUGGACCC | circ |
| ENSG00000100139:+:22:37912350:37932888 | ENSG00000100139 | MSTRG.22088.4 | + | 22 | 37912351 | 37932888 | 22 | CUCAUCUAUGUGCCUUUGAAGU | bsj |
| ENSG00000100139:+:22:37912350:37932888 | ENSG00000100139 | MSTRG.22088.4 | + | 22 | 37912151 | 37912360 | 210 | AGCCUCCUCAUGCAUGGAGCACCAGGCUUCCCCUUUCUCCAGGCCAUCCUGUGGUGCUUGGGUGGGAUUAAUACUGAGGGGAGCCCACAGUCACCCCAUCCCACUGAAGGGAGCUGGGCCUAGGCUGAGGGGUCGCCCCCUAACGCCUCCUCCUCUUCCUGCUUCGGCUGCUCCCGCCCUUGUACCUGUCACCACCCCAGGCCUUUGAAG | ie_up |
| ENSG00000100139:+:22:37912350:37932888 | ENSG00000100139 | MSTRG.22088.4 | + | 22 | 37932879 | 37933088 | 210 | UCAUCUAUGUGUGAGUCCCCCCGCCUGGGGCAUCCCUCCCUGGAAUCCGUAGAGCUUAGAAUGGAGAACCCUUACCCUCUUCCCUCAUCAGUAACAGCGCAGGGCAGGGCAGCCGGGGCUCGGGCAGAAUUGUUAAGAGUCACCUUAUUCCCACCCCUAGCUUCAAGCAGCAGAACCUGGAGCAGCGCCAGGCUGAUGUCGAGUAUGAGC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
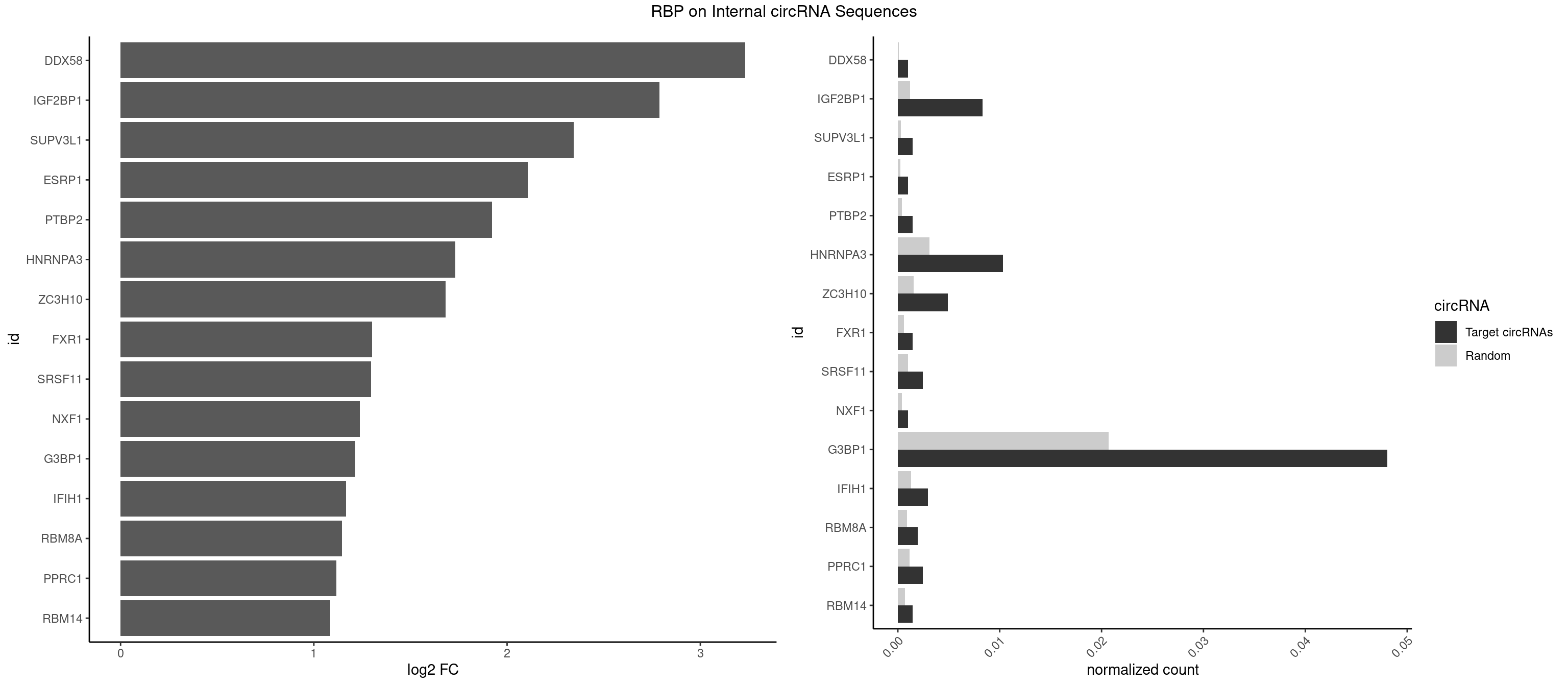
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.000980873 | 0.0001043427 | 3.232737 | GCGCGC | GCGCGC |
| IGF2BP1 | 16 | 831 | 0.008337420 | 0.0012057377 | 2.789685 | AAGCAC,AGCACC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SUPV3L1 | 2 | 199 | 0.001471309 | 0.0002898408 | 2.343768 | CCGCCC | CCGCCC |
| ESRP1 | 1 | 156 | 0.000980873 | 0.0002275250 | 2.108041 | AGGGAU | AGGGAU |
| PTBP2 | 2 | 267 | 0.001471309 | 0.0003883867 | 1.921535 | CUCUCU | CUCUCU |
| HNRNPA3 | 20 | 2140 | 0.010299166 | 0.0031027457 | 1.730910 | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ZC3H10 | 9 | 1053 | 0.004904365 | 0.0015274610 | 1.682931 | CAGCGC,CCAGCG,GAGCGC,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| FXR1 | 2 | 411 | 0.001471309 | 0.0005970720 | 1.301124 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 4 | 688 | 0.002452182 | 0.0009985015 | 1.296230 | AAGAAG | AAGAAG |
| NXF1 | 1 | 286 | 0.000980873 | 0.0004159215 | 1.237755 | AACCUG | AACCUG |
| G3BP1 | 97 | 14282 | 0.048062776 | 0.0206989801 | 1.215360 | ACCCAC,ACCCCC,ACCCCU,ACCGGC,ACGCCC,AGGCAC,AGGCAG,AGGCCC,AGGCCG,CACCCG,CACCGG,CACGCC,CAGGCA,CAGGCC,CCACAC,CCACAG,CCACCC,CCACGC,CCAGGC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCGC,CCCCGG,CCCCUA,CCCGCC,CCCGGC,CCCUAC,CCCUCG,CCGCAG,CCGCCC,CCGCCG,CCGGCC,CCUCGG,CGGCAG,CGGCCC,CGGCCG,CUCGGC,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| IFIH1 | 5 | 904 | 0.002942619 | 0.0013115296 | 1.165850 | GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 3 | 611 | 0.001961746 | 0.0008869128 | 1.145274 | CGCGCG,GCGCGC,GUGCGC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 4 | 780 | 0.002452182 | 0.0011318283 | 1.115411 | CCGCGC,CGCGCG,GCGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| RBM14 | 2 | 478 | 0.001471309 | 0.0006941687 | 1.083743 | CGCGCG,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
RBP on BSJ (Exon Only)
Plot
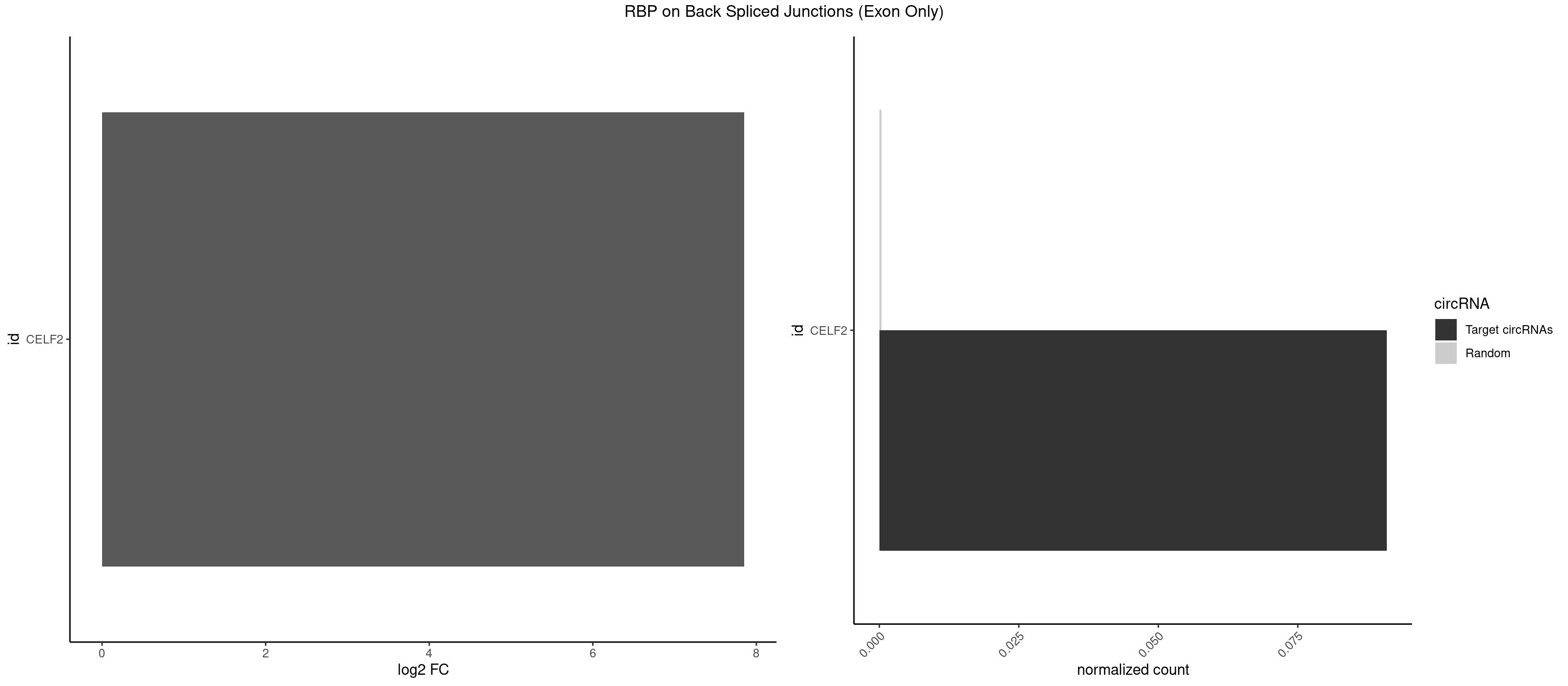
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF2 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | UAUGUG | AUGUGU,GUCUGU,UAUGUU,UGUGUG,UGUUGU |
RBP on BSJ (Exon and Intron)
Plot
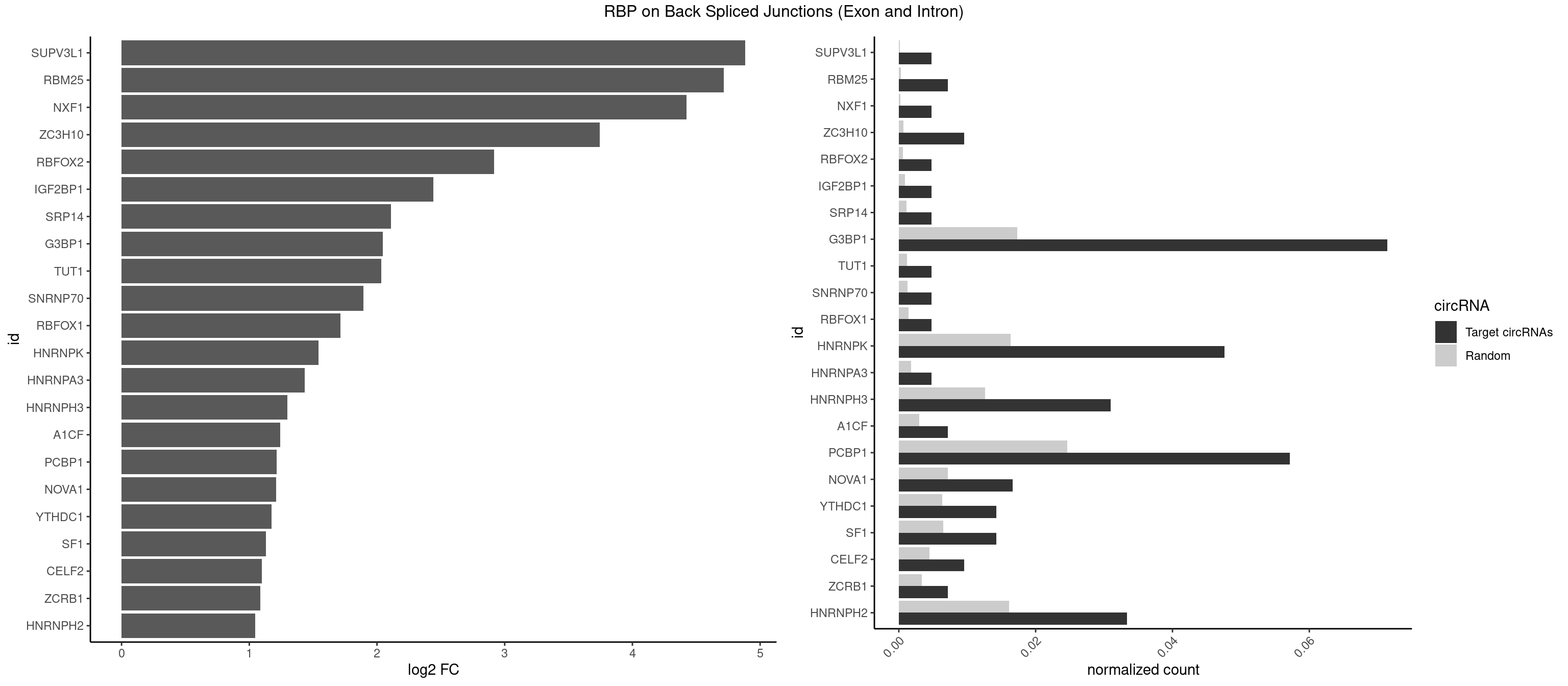
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 31 | 0.004761905 | 0.0001612253 | 4.884389 | CCGCCC | CCGCCC |
| RBM25 | 2 | 53 | 0.007142857 | 0.0002720677 | 4.714464 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| ZC3H10 | 3 | 140 | 0.009523810 | 0.0007103990 | 3.744837 | CAGCGC,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| G3BP1 | 29 | 3431 | 0.071428571 | 0.0172914148 | 2.046445 | ACCCCU,CAGGCC,CCACAG,CCACCC,CCAGGC,CCAUCC,CCCACA,CCCACC,CCCAGG,CCCAUC,CCCCAG,CCCCCC,CCCCCG,CCCCGC,CCCCUA,CCCGCC,CCCUAG,CCCUCC,CCGCCC,CCUAGG,CUAGGC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPK | 19 | 3244 | 0.047619048 | 0.0163492543 | 1.542314 | ACCCCA,ACCCUU,CAUCCC,CCAUCC,CCCCAU,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAC,GCCCCC,UCCCAC,UCCCCC,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPH3 | 12 | 2496 | 0.030952381 | 0.0125806127 | 1.298848 | AAGGGA,AGGGGA,GAAGGG,GAGGGG,GGGGAG,GGGGCU,GGGUCG,GUCGAG,UCGGGC,UGGGUG,UUGGGU | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUCAGU,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PCBP1 | 23 | 4891 | 0.057142857 | 0.0246473196 | 1.213142 | ACCCUC,ACCUUA,AUUCCC,CAUCCC,CCACCC,CCAUCC,CCCACC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCUAAC,CCUCCC,CCUCUU,CCUUAC,CCUUUC,CUUACC,CUUCCC | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | ACCACC,AGCACC,AGUCAC,CAGUCA,CCCCCC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GGCUGC,UAAUAC,UCAUGC,UCCUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SF1 | 5 | 1294 | 0.014285714 | 0.0065245869 | 1.130615 | ACAGUC,AGUAAC,CACUGA,CAGUCA,UACUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,UAUGUG,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPH2 | 13 | 3198 | 0.033333333 | 0.0161174929 | 1.048338 | AAGGGA,AGGGGA,CUGGGG,GAAGGG,GAGGGG,GGGGAG,GGGGCU,GGGUCG,GUCGAG,UCGGGC,UGGGUG,UUGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.