circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000229807:-:X:73826114:73833374
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000229807:-:X:73826114:73833374 | ENSG00000229807 | ENST00000650627 | - | X | 73826115 | 73833374 | 2380 | GACAUUCUGAGCAUGUGAGACCUGAGGACUGCAAACAGCUAUAAGAGGCUCCAAAUUAAUCAUAUCUUUCCCUUUGAGAAUCUGGCCAAGCUCCAGCUAAUCUACUUGGAUGGGUUGCCAGCUAUCUGGAGAAAAAGAUCUUCCUCAGAAGAAUAGGCUUGUUGUUUUACAGUGUUAGUGAUCCAUUCCCUUUGACGAUCCCUAGGUGGAGAUGGGGCAUGAGGAUCCUCCAGGGGAAAAGCUCACUACCACUGGGCAACAACCCUAGGUCAGGAGGUUCUGUCAAGAUACUUUCCUGGUCCCAGAUAGGAAGAUAAAGUCUCAAAAACAACCACCACACGUCAAGCUCUUCAUUGUUCCUAUCUGCCAAAUCAUUAUACUUCCUACAAGCAGUGCAGAGAGCUGAGUCUUCAGCAGGUCCAAGAAAUUUGAACACACUGAAGGAAGUCAGCCUUCCCACCUGAAGAUCAACAUGCCUGGCACUCUAGCACUUGAGGAUAGCUGAAUGAAUGUGUAUUUCUUUGUCUCUUUCUUUCUUGUCUUUGCUCUUUGUUCUCUAUCUAAAGUGUGUCUUACCCAUUUCCAUGUUUCUCUUGCUAAUUUCUUUCGUGUGUGCCUUUGCCUCAUUUUCUCUUUUUGUUCACAAGAGUGGUCUGUGUCUUGUCUUAGACAUAUCUCUCAUUUUUCAUUUUGUUGCUAUUUCUCUUUGCUCUCCUAGAUGUGGCUCUUCUUUCACGCUUUAUUUCAUGUCUCCUUUUUGGGUCACAUGCUGUGUGCUUUUUGUCCUUUUCUUGUUCUGUCUACCUCUCCUUUCUCUGCCUACCUCUCUUUUCUCUUUGUGAACUGUGAUUAUUUGUUACCCCUUCCCCUUCUCGUUCGUUUUAAAUUUCACCUUUUUUCUGAGUCUGGCCUCCUUUCUGCUGUUUCUACUUUUUAUCUCACAUUUCUCAUUUCUGCAUUUCCUUUCUGCCUCUCUUGGGCUAUUCUCUCUCUCCUCCCCUGCGUGCCUCAGCAUCUCUUGCUGUUUGUGAUUUUCUAUUUCAGUAUUAAUCUCUGUUGGCUUGUAUUUGUUCUCUGCUUCUUCCCUUUCUACUCACCUUUGAGUAUUUCAGCCUCUUCAUGAAUCUAUCUCCCUCUCUUUGAUUUCAUGUAAUCUCUCCUUAAAUAUUUCUUUGCAUAUGUGGGCAAGUGUACGUGUGUGUGUGUCAUGUGUGGCAGAGGGGCUUCCUAACCCCUGCCUGAUAGGUGCAGAACGUCGGCUAUCAGAGCAAGCAUUGUGGAGCGGUUCCUUAUGCCAGGCUGCCAUGUGAGAUGAUCCAAGACCAAAACAAGGCCCUAGACUGCAGUAAAACCCAGAACUCAAGUAGGGCAGAAGGUGGAAGGCUCAUAUGGAUAGAAGGCCCAAAGUAUAAGACAGAUGGUUUGAGACUUGAGACCCGAGGACUAAGAUGGAAAGCCCAUGUUCCAAGAUAGAUAGAAGCCUCAGGCCUGAAACCAACAAAAGCCUCAAGAGCCAAGAAAACAGAGGGUGGCCUGAAUUGGACCGAAGGCCUGAGUUGGAUGGAAGUCUCAAGGCUUGAGUUAGAAGUCUUAAGACCUGGGACAGGACACAUGGAAGGCCUAAGAACUGAGACUUGUGACACAAGGCCAACGACCUAAGAUUAGCCCAGGGUUGUAGCUGGAAGACCUACAACCCAAGGAUGGAAGGCCCCUGUCACAAAGCCUACCUAGAUGGAUAGAGGACCCAAGCGAAAAAGGUAUCUCAAGACUAACGGCCGGAAUCUGGAGGCCCAUGACCCAGAACCCAGGAAGGAUAGAAGCUUGAAGACCUGGGGAAAUCCCAAGAUGAGAACCCUAAACCCUACCUCUUUUCUAUUGUUUACACUUCUUACUCUUAGAUAUUUCCAGUUCUCCUGUUUAUCUUUAAGCCUGAUUCUUUUGAGAUGUACUUUUUGAUGUUGCCGGUUACCUUUAGAUUGACAGUAUUAUGCCUGGGCCAGUCUUGAGCCAGCUUUAAAUCACAGCUUUUACCUAUUUGUUAGGCUAUAGUGUUUUGUAAACUUCUGUUUCUAUUCACAUCUUCUCCACUUGAGAGAGACACCAAAAUCCAGUCAGUAUCUAAUCUGGCUUUUGUUAACUUCCCUCAGGAGCAGACAUUCAUAUAGGUGAUACUGUAUUUCAGUCCUUUCUUUUGACCCCAGAAGCCCUAGACUGAGAAGAUAAAAUGGUCAGGUUGUUGGGGAAAAAAAAGUGCCAGGCUCUCUAGAGAAAAAUGUGAAGAGAUGCUCCAGGCCAAUGAGAAGAAUUAGACAAGAAAUACACAGAUGUGCCAGACUUCUGAGAAGCACCUGCCAGCAACAGCUUCCUUCUUUGAGCUUAGGACAUUCUGAGCAUGUGAGACCUGAGGACUGCAAACAGCUAUAAGAGGCU | circ |
| ENSG00000229807:-:X:73826114:73833374 | ENSG00000229807 | ENST00000650627 | - | X | 73826115 | 73833374 | 22 | UUUGAGCUUAGGACAUUCUGAG | bsj |
| ENSG00000229807:-:X:73826114:73833374 | ENSG00000229807 | ENST00000650627 | - | X | 73833365 | 73833574 | 210 | CUAAUCUUCCCGUGUACUGCCAGGGCUUGUCAUUAGAGGACUUUAGGGAGACCAAGCAGGCUAGAAAGUAGAGACAGGAGAUACCUAUGUCUAAUGCUUCAGUUUAUACUUCCUAGGUUUUUUUCAUUGGGGUUUUUGUAACUCUUUUGGUAUCCUACCGGUGCUUUGGUAGCCUACUGAACCCUGUCUUUCUUCUUAAGGACAUUCUGA | ie_up |
| ENSG00000229807:-:X:73826114:73833374 | ENSG00000229807 | ENST00000650627 | - | X | 73825915 | 73826124 | 210 | UUGAGCUUAGGUGAGCAGGAUUCUGGGGUUUGGGAUUUCUAGUGAUGGUUAUGGAAAGGGUGACUGUGCCUGGGACAAAGCGAGGUCCCAAGGGGACAGCCUGAACUCCCUGCUCAUAGUAGUGGCCAAAUAAUUUGGUGGACUGUGCCAACGCUACUCCUGGGUUUAAUACCCAUCUCUAGGCUUAAAGAUGAGAGAACCUGGGACUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
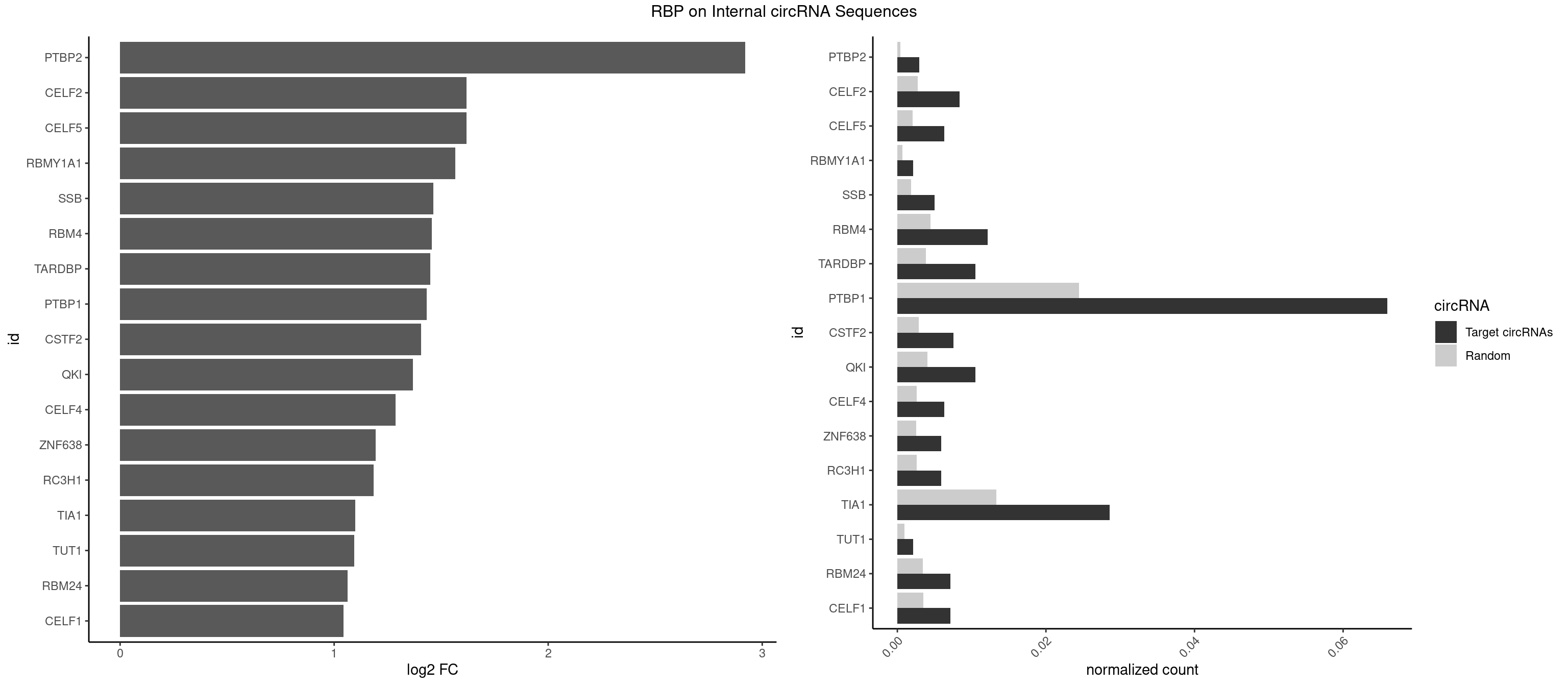
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 6 | 267 | 0.002941176 | 0.0003883867 | 2.920828 | CUCUCU | CUCUCU |
| CELF2 | 19 | 1886 | 0.008403361 | 0.0027346479 | 1.619611 | AUGUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 14 | 1415 | 0.006302521 | 0.0020520728 | 1.618847 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBMY1A1 | 4 | 489 | 0.002100840 | 0.0007101099 | 1.564852 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| SSB | 11 | 1260 | 0.005042017 | 0.0018274462 | 1.464172 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM4 | 28 | 3065 | 0.012184874 | 0.0044432593 | 1.455401 | CCUCUU,CCUUCC,CCUUCU,CUCUUU,CUUCCU,CUUCUU,UCCUUC,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| TARDBP | 24 | 2654 | 0.010504202 | 0.0038476365 | 1.448922 | GAAUGA,GAAUGU,GUGUGU,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PTBP1 | 156 | 16854 | 0.065966387 | 0.0244263326 | 1.433294 | ACUUUC,ACUUUU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUGUG,GGCUCC,GUCUUA,GUCUUU,UACUUU,UAGCUG,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| CSTF2 | 17 | 1967 | 0.007563025 | 0.0028520334 | 1.406973 | GUGUGU,GUGUUU,GUUUUG,UGUGUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| QKI | 24 | 2805 | 0.010504202 | 0.0040664663 | 1.369119 | AAUCAU,ACACAC,ACUAAC,AUCAUA,AUCUAA,AUCUAC,CACACU,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CELF4 | 14 | 1782 | 0.006302521 | 0.0025839306 | 1.286362 | GUGUGG,GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ZNF638 | 13 | 1773 | 0.005882353 | 0.0025708878 | 1.194127 | CGUUCG,GGUUCU,GGUUGU,GUUCGU,GUUGUU,UGUUCU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 13 | 1786 | 0.005882353 | 0.0025897275 | 1.183593 | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| TIA1 | 67 | 9206 | 0.028571429 | 0.0133428208 | 1.098509 | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUG,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TUT1 | 4 | 678 | 0.002100840 | 0.0009840095 | 1.094222 | AAAUAC,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM24 | 16 | 2357 | 0.007142857 | 0.0034172229 | 1.063677 | AGAGUG,AGUGUG,GAGUGG,GUGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 | 16 | 2391 | 0.007142857 | 0.0034664959 | 1.043023 | CUGUCU,GUGUGU,GUUUGU,UGUGUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
RBP on BSJ (Exon Only)
Plot
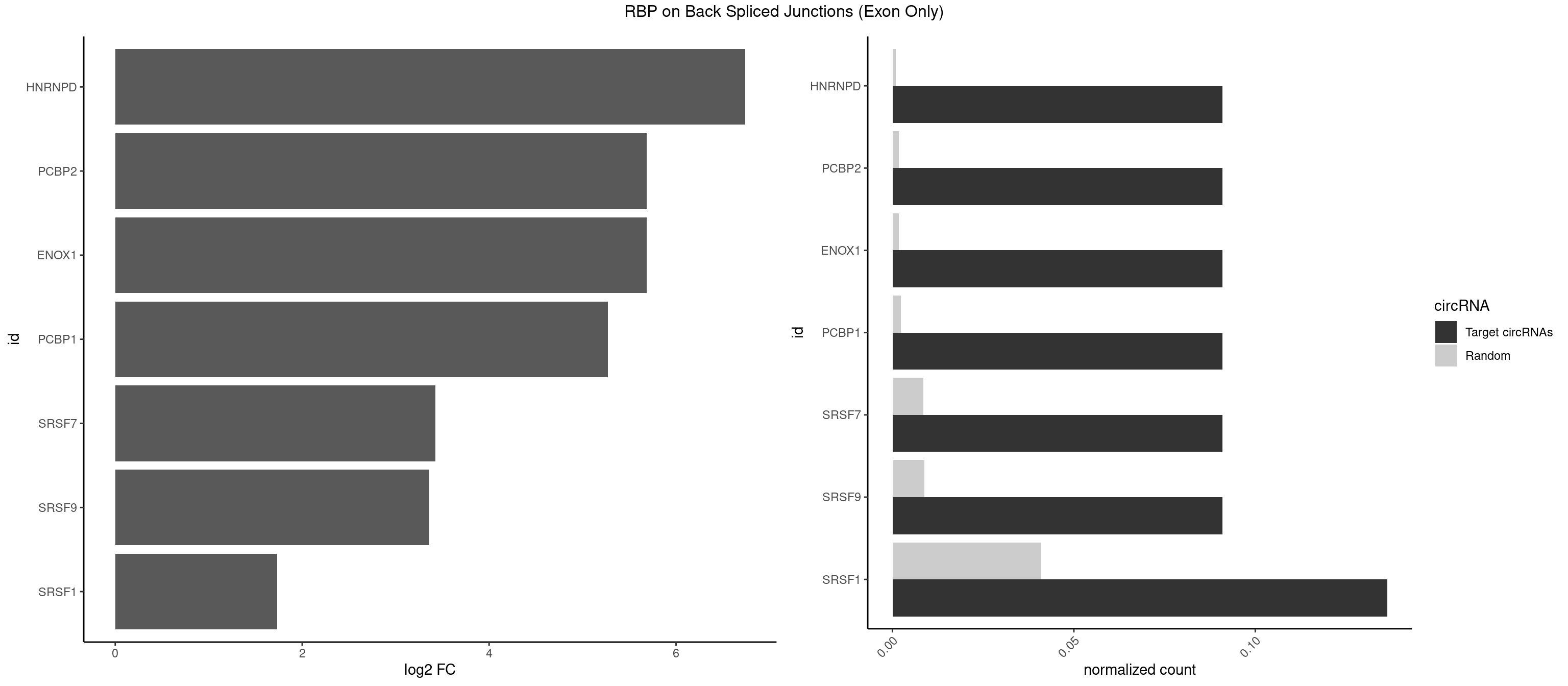
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPD | 1 | 12 | 0.09090909 | 0.000851454 | 6.738352 | UUAGGA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,UUAGAG,UUAGGA,UUAGGG |
| ENOX1 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | AGGACA | AAGACA,AGACAG,AGGACA,AGUACA,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,UAGACA,UGGACA |
| PCBP2 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | UUAGGA | AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,AUUAAA,CCAUUC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CUCCCU,CUUCCA,UAACCC,UUAGAG,UUAGGA,UUAGGG |
| PCBP1 | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | UUAGGA | AAAAAA,AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,ACCCUC,AUUAAA,CAAACC,CAUACC,CCAACC,CCACCC,CCAUAC,CCAUUC,CCCACC,CCCCAC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,UUAGAG,UUAGGA,UUAGGG |
| SRSF7 | 1 | 128 | 0.09090909 | 0.008449044 | 3.427565 | AGGACA | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
| SRSF9 | 1 | 134 | 0.09090909 | 0.008842023 | 3.361976 | AGGACA | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SRSF1 | 2 | 625 | 0.13636364 | 0.041000786 | 1.733736 | AGGACA,UAGGAC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
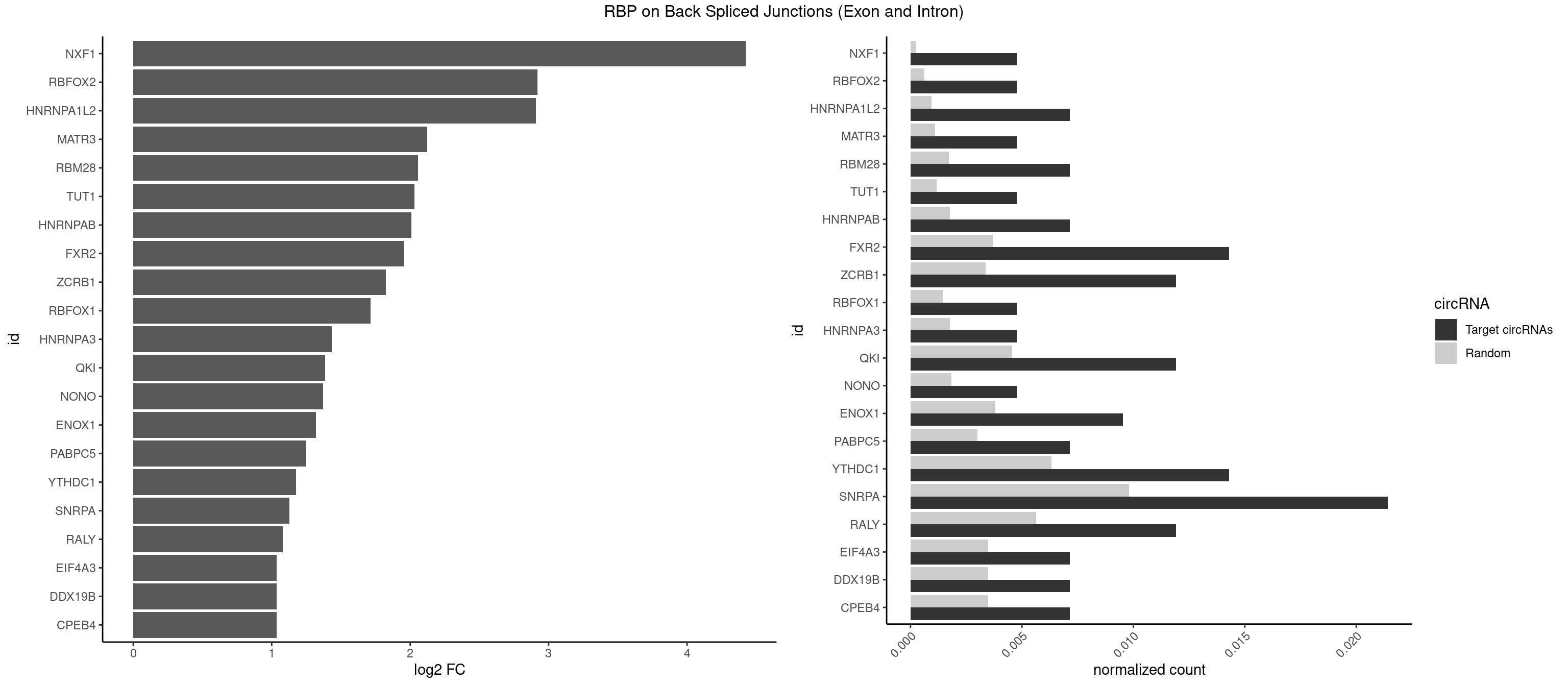
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGA,AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | ACAAAG,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| FXR2 | 5 | 730 | 0.014285714 | 0.0036829907 | 1.955624 | AGACAG,GACAAA,GACAGG,GGACAA,GGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | GCUUAA,GGCUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | CUAAUC,CUACUC,CUCAUA,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGACAG,AGGACA,GGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GCCUAC,UAAUAC,UAAUGC,UACUGC,UCCUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| SNRPA | 8 | 1947 | 0.021428571 | 0.0098145909 | 1.126536 | AGGAGA,AGUAGU,AUACCU,CCUGCU,GAGCAG,GAUACC,GGAGAU,GUAGUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.