circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000133612:+:7:151116792:151134568
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000133612:+:7:151116792:151134568 | ENSG00000133612 | MSTRG.30937.6 | + | 7 | 151116793 | 151134568 | 2459 | ACUCGUUUGUGAACAGCCAGGAGUGGACGCUGAGCCGCUCCGUACCGGAGCUUAAAGUGGGCAUAGUGGGGAACCUGUCUAGCGGGAAGUCAGCCCUGGUGCACCGCUAUCUGACGGGGACCUAUGUCCAGGAGGAGUCCCCUGAAGGGGGGCGGUUUAAGAAGGAGAUUGUGGUGGAUGGCCAGAGUUACCUGCUGCUGAUCCGAGAUGAAGGAGGCCCCCCUGAGCUCCAGUUUGCUGCCUGGGUGGAUGCAGUGGUGUUUGUGUUCAGCCUGGAGGAUGAAAUCAGUUUCCAGACGGUGUACAACUACUUCCUGCGUCUCUGCAGCUUCCGCAACGCCAGCGAGGUGCCCAUGGUGCUUGUGGGCACGCAGGAUGCCAUCAGCGCUGCGAAUCCCCGGGUUAUCGACGACAGCAGAGCCCGCAAGCUCUCCACAGAUCUGAAGCGGUGCACCUACUAUGAGACGUGCGCGACCUACGGGCUCAAUGUGGAGCGUGUCUUCCAGGACGUGGCCCAGAAGGUAGUGGCCUUGCGAAAGAAGCAGCAACUGGCCAUCGGGCCCUGCAAGUCACUGCCCAACUCGCCCAGCCACUCGGCCGUGUCCGCCGCCUCCAUCCCGGCCGUGCACAUCAACCAGGCCACGAAUGGCGGCGGCAGCGCCUUCAGCGACUACUCGUCCUCAGUCCCCUCCACCCCCAGCAUCAGCCAGCGGGAGCUGCGCAUCGAGACCAUCGCUGCCUCCUCCACCCCCACACCCAUCCGAAAGCAGUCCAAGCGGCGCUCCAACAUCUUCACGCUAAUGAACCGGCGGCCGGAAGGCUGUUUGCCCAAGGCGACACAGAGGGUUAGCUGGCCCAGCUAGAGUCAGAGCCCAGGCCUCCCCGUCGCCUUCCGGCUCUUUUUCCGUUACCCCACUGCCGAGGGGAAAAUUCAUGCUUUCCACCCACCGCUGUGAUUGGUUUAACAAAGGCUUAGCCAUCUUUUCCUGGGCUUCACCACGCUGCCUCGUUCCUCCCUCUCAGAUAUGUGCCACUGUUUCCAACUUUUCAUCAACAAAAAGGCCUUUCCAACUCCUUCCAAAUUAGAAGACCAGUUGGUGACACACAGCACCUCUGGACAUGCCCCCUGUGCGGGGCCGGAGGCGGGCCGGGCCCUGGGACUGCUCUCAGAUGAGAAGCGGCCGCCGAGCUCCCCACUCCAGAGACCCACGGGAACCUUUGUAACUAACCCCACCCCCAGGGAAGGCUAGGAGCGCCGGAGCCCGCGCUGGGGGCUGCCGGGGACCAGGCCCGGCCGGACGCUGCAGGCUCGCUGCAUGGAGAAGAAGGCAGCUCGGCCCCACGCCCGGCGCUGGCCAGCGCGACGAGGCCCAGAGGGGCGGGGGAGUCCAAGCCCGCCCGGCCCGGCUGCUCCUGGGGGGCGCUUUCCUGCCCCUCCCCUCCUCUGCUCCUUCCUGCAUGGACCUCUGCCGUUCCUGCUCCUGCUCCGGAAGCCGCCGCCGCCGCCGCGCUUGCCUUGCCCCCUCUUUUUGGCCUCCCCCUAUUUCCUAGGAUCCGCAUUCGGGUGGACUCUGCCCCAGGAGCUUGACAGGGUGCAGGGCCUCUUCUGGCCUCUCUCCUCUCUCUGUCCAUCCACUGUGCCCCUUGGGCCACGCCGACGCGCUCGGUGCCACGUGCCGUGUGGUGUCUGUGCCGCAGACGGGCCAUCUGCCCGCUCACUUGUGGACUUUGCGCUCCCGGUUGUCGCGCCCUGUGCUCCCGACCAGCAGCCCCACCGUCCACCUCCUCGCCCCCGCUUCUCUUACGCCCCCGCCCCGGGCGUGCUCCUCGCGCCCUCGGCCUCUCUGUCCUUGCUCUUUUGUAAGGGGCGGGCCCGGCUCAGUGACCUGUGCUGCUCUGUAUGGUGCCGUGUGUAAGAAUAAACCCGUUGGAAUACUCGUGGUCUCAGUUCCUUCCCGUCCCGCCGCCCCGGCCCGACCCACUGCUAGGGCUGCCCAGGAGGAGGGAGGCAGGAGGGAGGCCGGGAAGUCCAGGUGGGCAGCUCUCGGCAGACCCUGGGCCUCUUUAACACGCCUCUUGUUUUCUCUUCCAGUCUCGGAAGGGUGCUGACCUGGACCGGGAGAAGAAGGCUGCCGAGUGCAAGGUGGACAGCAUCGGGAGCGGCCGCGCCAUCCCCAUCAAGCAGGGGAUCCUGCUAAAGCGGAGCGGCAAGUCCCUGAACAAGGAGUGGAAGAAGAAGUAUGUGACGCUCUGUGACAACGGGCUGCUCACCUAUCACCCCAGCCUGCAUGAUUACAUGCAGAACAUCCACGGCAAGGAGAUUGACCUGCUGCGGACAACGGUGAAAGUGCCAGGGAAGCGCCUGCCCCGAGCCACACCUGCCACAGCCCCGGGCACCAGCCCCCGUGCCAACGGGCUGUCCGUGGAGCGGAGUAACACACAGCUGGGUGGGGGCACAGACUCGUUUGUGAACAGCCAGGAGUGGACGCUGAGCCGCUCCGUACCGGAG | circ |
| ENSG00000133612:+:7:151116792:151134568 | ENSG00000133612 | MSTRG.30937.6 | + | 7 | 151116793 | 151134568 | 22 | UGGGGGCACAGACUCGUUUGUG | bsj |
| ENSG00000133612:+:7:151116792:151134568 | ENSG00000133612 | MSTRG.30937.6 | + | 7 | 151116593 | 151116802 | 210 | CAUCCUUACUCUGCUUCACUCAAGAAGAACCUUCCCACUGCCUCCUGGGGAACUAGGGGUGAGGGUUGGGGGUCCCGUGGAGGAAGCUGCUGUCCUCUGGCCUGGGCUUGGGGAGGGCAUCAUAGGUGACACAGGCAGCAGUGGUUGGGACAGGUGUGUGUGCCACCCUGGCCCUGACGGGGCGGCUCUGUCUUCCGCAGACUCGUUUGU | ie_up |
| ENSG00000133612:+:7:151116792:151134568 | ENSG00000133612 | MSTRG.30937.6 | + | 7 | 151134559 | 151134768 | 210 | GGGGGCACAGGUGAGGCGGCUGCUGAGGUGGGGGCCUGGGGGGUGGCUGCCUUGGAGCCAAGGCAAGCAGGCAUUCUGGGCUUGGCUGCUUCUCAGCCUGGGCACAGGGUGCUGGCCAGCAUCACACUUCAAGGUCAUCUCAGCCAAAGACAUGUGCCCUAACGCAGCCCUCGUGGAGUUAGUUGAGGUGACAGCAAAUGCCUAGACGAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
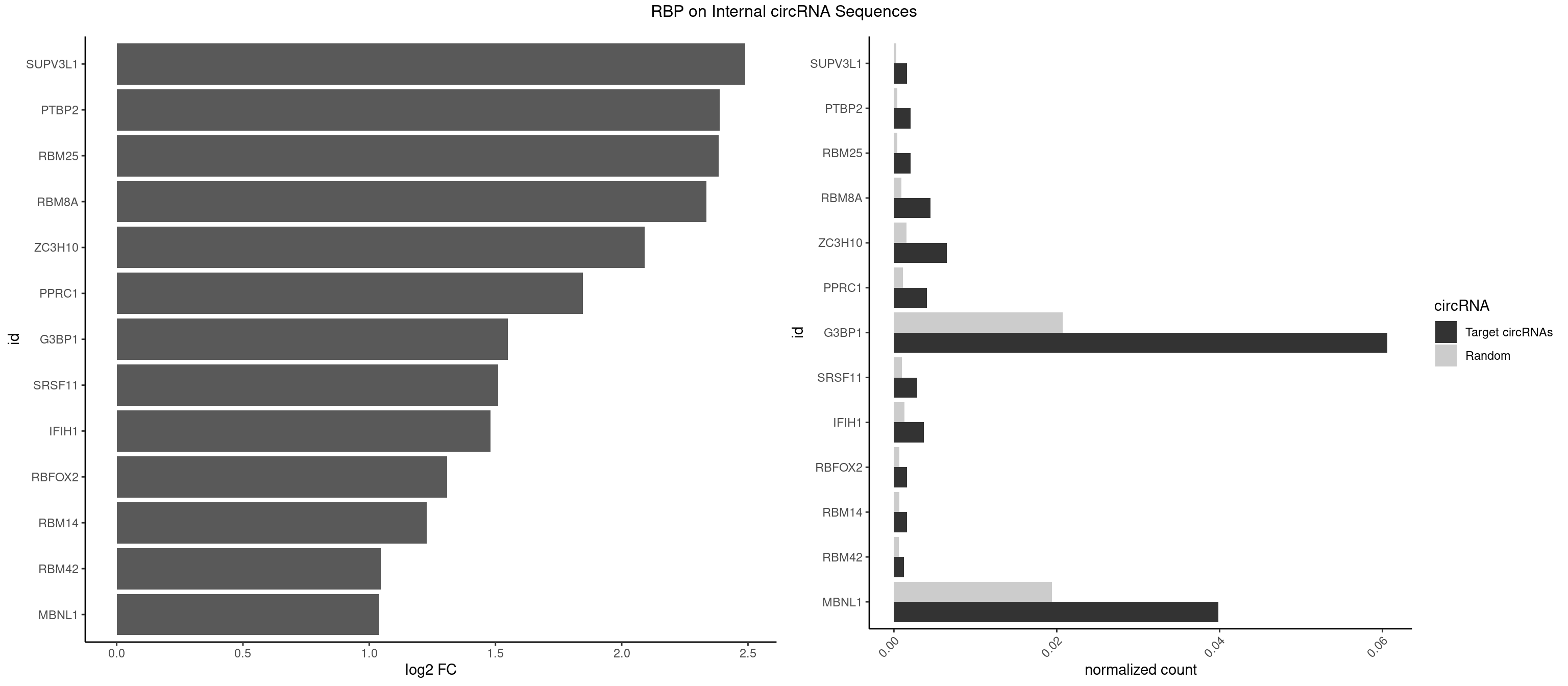
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 3 | 199 | 0.001626678 | 0.0002898408 | 2.488596 | CCGCCC | CCGCCC |
| PTBP2 | 4 | 267 | 0.002033347 | 0.0003883867 | 2.388291 | CUCUCU | CUCUCU |
| RBM25 | 4 | 268 | 0.002033347 | 0.0003898359 | 2.382918 | AUCGGG,CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| RBM8A | 10 | 611 | 0.004473363 | 0.0008869128 | 2.334496 | ACGCGC,CGCGCC,CGCGCU,GUGCGC,UGCGCG,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| ZC3H10 | 15 | 1053 | 0.006506710 | 0.0015274610 | 2.090793 | CAGCGA,CAGCGC,CCAGCG,GAGCGC,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PPRC1 | 9 | 780 | 0.004066694 | 0.0011318283 | 1.845201 | CCGCGC,CGCGCC,CGGCGC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| G3BP1 | 148 | 14282 | 0.060593737 | 0.0206989801 | 1.549609 | ACACGC,ACCCAC,ACCCAU,ACCCCC,ACCGGC,ACGCAG,ACGCCC,ACGCCG,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACGCA,CACGCC,CAGGCC,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUCC,CCCUCG,CCGCAG,CCGCCC,CCGCCG,CCGGCC,CCUACG,CCUAGG,CCUCGG,CGGCAG,CGGCCC,CGGCCG,CUCGGC,UACGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| SRSF11 | 6 | 688 | 0.002846686 | 0.0009985015 | 1.511447 | AAGAAG | AAGAAG |
| IFIH1 | 8 | 904 | 0.003660024 | 0.0013115296 | 1.480603 | GCCGCG,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBFOX2 | 3 | 452 | 0.001626678 | 0.0006564894 | 1.309085 | UGCAUG | UGACUG,UGCAUG |
| RBM14 | 3 | 478 | 0.001626678 | 0.0006941687 | 1.228570 | CGCGCC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM42 | 2 | 407 | 0.001220008 | 0.0005912752 | 1.044989 | AACUAA,AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| MBNL1 | 97 | 13372 | 0.039853599 | 0.0193802045 | 1.040126 | ACGCUA,ACGCUC,ACGCUG,AUGCCA,AUGCCC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GUCUCG,GUGCUC,GUGCUG,GUGCUU,UCGCUG,UCUGCU,UGCUGC,UGCUUU,UUGCCC,UUGCCU,UUGCUC,UUGCUG,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
RBP on BSJ (Exon Only)
Plot
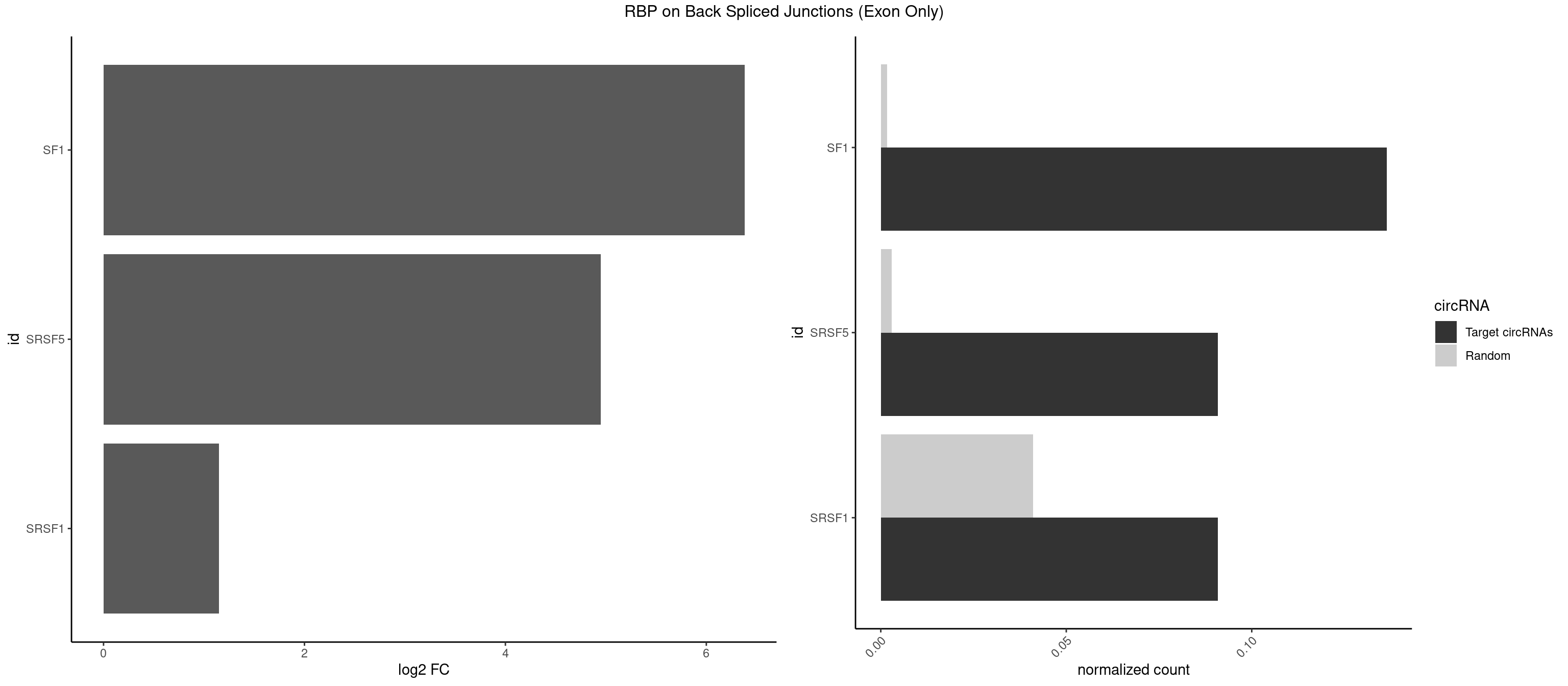
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SF1 | 2 | 24 | 0.13636364 | 0.001637412 | 6.379898 | ACAGAC,CACAGA | ACAGAC,ACAGUC,AGUAAG,AUACUA,CACAGA,CACUGA,CAGUCA,GCUGAC,GCUGCC,UACUGA,UGCUAA,UGCUGA,UGCUGC |
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | CACAGA | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| SRSF1 | 1 | 625 | 0.09090909 | 0.041000786 | 1.148773 | CACAGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
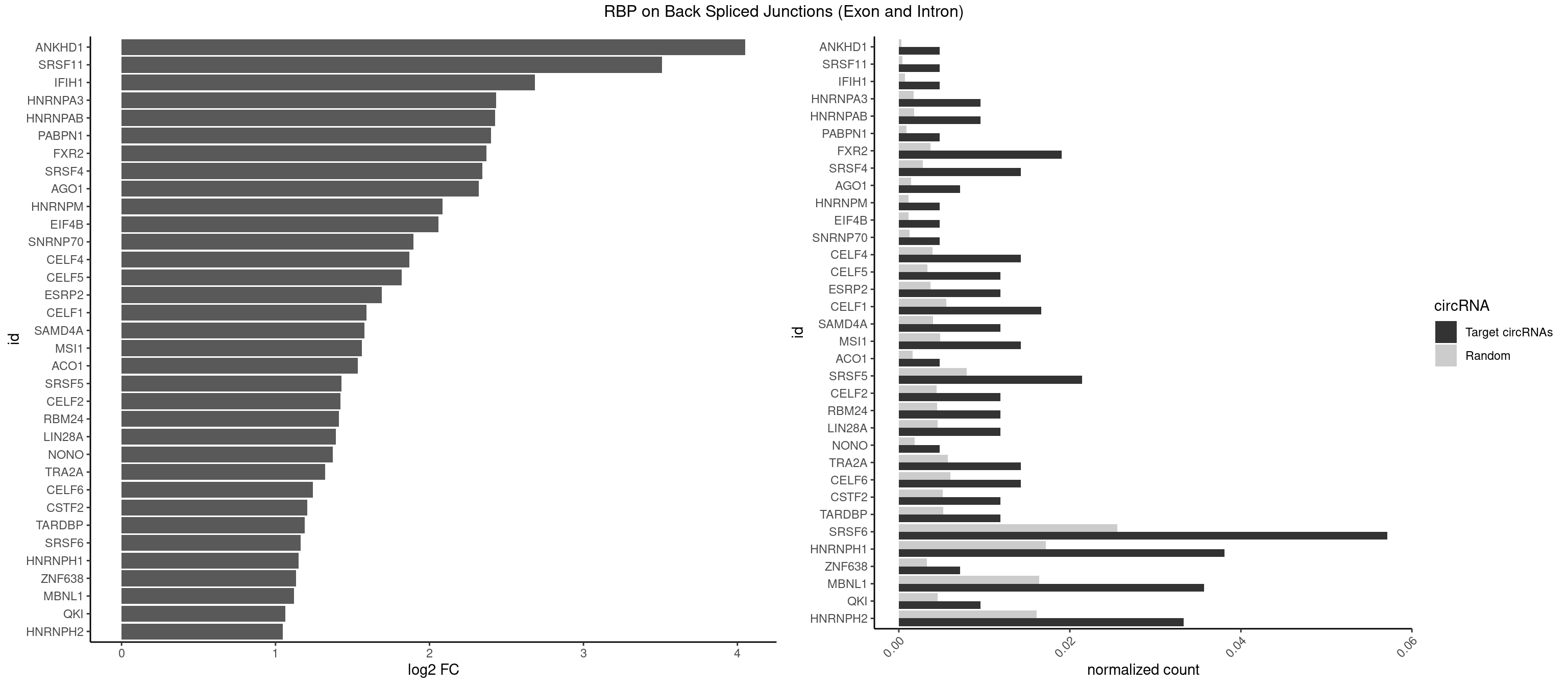
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPA3 | 3 | 349 | 0.009523810 | 0.0017634019 | 2.433177 | CCAAGG,GCCAAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 3 | 351 | 0.009523810 | 0.0017734784 | 2.424957 | AAAGAC,AAGACA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| FXR2 | 7 | 730 | 0.019047619 | 0.0036829907 | 2.370661 | AGACGA,GACAGG,GACGAG,GACGGG,GGACAG,UGACAG,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRSF4 | 5 | 558 | 0.014285714 | 0.0028164047 | 2.342647 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GGUGUG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ESRP2 | 4 | 731 | 0.011904762 | 0.0036880290 | 1.690617 | GGGGAA,GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | CUGUCU,GUGUGU,GUUUGU,UGUGUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGGAAG,AGGUGG,AGUUAG,UAGGUG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRSF5 | 8 | 1578 | 0.021428571 | 0.0079554615 | 1.429518 | AAGAAG,AGAAGA,AGGAAG,CCGCAG,CGCAGA,CGCAGC,GAAGAA,GAGGAA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUGUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGGA,GGAGGG,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGAGG,GUGGGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF6 | 23 | 5069 | 0.057142857 | 0.0255441354 | 1.161581 | AAGAAG,AGAAGA,AGGAAG,CACAGG,CAUCCU,CCACUG,CCUCUG,CCUGGC,CUCUGG,CUUCAC,CUUCUC,GAAGAA,GAGGAA,GCAGCA,UCACAC,UCACUC,UCCUGG,UCUCAG,UUACUC,UUCUGG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| HNRNPH1 | 15 | 3406 | 0.038095238 | 0.0171654575 | 1.150102 | AGGAAG,CUGGGG,GAGGAA,GGAGGA,GGAGGG,GGGAGG,GGGGAA,GGGGAG,GGGGGC,GGGGGG,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| MBNL1 | 14 | 3258 | 0.035714286 | 0.0164197904 | 1.121066 | CUGCCU,CUGCUG,CUGCUU,GCUGCU,GUGCUG,UCUGCU,UGCUGU,UGCUUC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | AUCAUA,CACACU,CUAACG | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPH2 | 13 | 3198 | 0.033333333 | 0.0161174929 | 1.048338 | CUGGGG,GGAGGA,GGAGGG,GGGAGG,GGGGAA,GGGGAG,GGGGGC,GGGGGG,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.