circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000146555:+:7:4049347:4114274
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000146555:+:7:4049347:4114274 | ENSG00000146555 | ENST00000404826 | + | 7 | 4049348 | 4114274 | 1221 | UGCCCACCGCGCCCCCGCAGAACGUGCAGACGGAAGCCGUGAACUCCACCACCAUUCAGUUCCUGUGGAACCCUCCGCCUCAGCAGUUUAUCAAUGGCAUCAACCAGGGAUACAAGCUUCUGGCAUGGCCGGCAGAUGCCCCCGAGGCUGUCACUGUGGUCACUAUUGCCCCAGAUUUCCACGGAGUCCACCAUGGACACAUAACGAACCUGAAGAAGUUUACCGCCUACUUCACUUCCGUUCUGUGCUUCACCACCCCUGGGGACGGGCCUCCCAGCACACCUCAGCUGGUCUGGACUCAGGAAGACAAACCAGGAGCUGUGGGACAUCUGAGUUUCACAGAGAUCUUGGACACAUCUCUCAAGGUCAGCUGGCAGGAGCCCCUGGAGAAAAAUGGCAUCAUUACUGGCUAUCAGAUCUCUUGGGAAGUGUACGGCAGGAACGACUCUCGUCUCACGCACACCCUGAACAGCACGACGCACGAGUACAAGAUCCAAGGCCUCUCAUCUCUCACCACCUACACCAUCGACGUGGCCGCUGUGACUGCCGUGGGCACUGGCCUGGUGACUUCAUCCACCAUUUCUUCUGGAGUGCCCCCAGACCUUCCUGGUGCCCCAUCCAACCUGGUCAUUUCCAACAUCAGCCCUCGCUCCGCCACCCUUCAGUUCCGGCCAGGCUAUGACGGGAAAACGUCCAUCUCCAGGUGGAUUGUUGAGGGGCAGGUGGGAGCUAUCGGCGACGAGGAGGAGUGGGUCACCCUCUAUGAAGAGGAGAAUGAGCCUGAUGCCCAGAUGCUGGAGAUCCCAAACCUCACACCCUACACUCACUACAGAUUUCGAAUGAAGCAAGUGAACAUUGUUGGGCCGAGCCCCUACAGUCCGUCUUCCCGGGUCAUCCAGACCCUGCAGGCCCCACCCGACGUGGCUCCAACCAGCGUCACGGUCCGUACUGCCAGUGAGACCAGCCUGCGGCUUCGCUGGGUGCCCCUGCCGGAUUCUCAGUACAACGGGAACCCCGAGUCCGUGGGCUACAGGAUUAAGUACUGGCGCUCAGACCUCCAGUCCUCAGCAGUGGCCCAAGUCGUCAGUGACCGGCUGGAGAGAGAAUUCACCAUCGAGGAGCUGGAGGAGUGGAUGGAAUACGAGCUGCAGAUGCAGGCCUUCAACGCCGUCGGGGCUGGGCCGUGGAGCGAGGUGGUGCGGGGCCGGACGCGGGAGUCAGUGCCCACCGCGCCCCCGCAGAACGUGCAGACGGAAGCCGUGAACUCCACC | circ |
| ENSG00000146555:+:7:4049347:4114274 | ENSG00000146555 | ENST00000404826 | + | 7 | 4049348 | 4114274 | 22 | GCGGGAGUCAGUGCCCACCGCG | bsj |
| ENSG00000146555:+:7:4049347:4114274 | ENSG00000146555 | ENST00000404826 | + | 7 | 4049148 | 4049357 | 210 | AAGAGAAAAUGAAAAAUAGCCCAAUUCUCUUUCUUGGUAAUGCCACUGUAAGCUUCCCAAAACAUUCAUCACACUUCCUCGGUCUCAGUGCAAUAAUUGCCUGUGAGCAUGAUGGCAGCUAAGGGUUUCCUUUCUGUCUCUCCACCCCAUGGUCCCUCCUGCCAUUUGUUCUGCUGUCAUCAAUGACUGCCCUGCCACAGUGCCCACCGC | ie_up |
| ENSG00000146555:+:7:4049347:4114274 | ENSG00000146555 | ENST00000404826 | + | 7 | 4114265 | 4114474 | 210 | CGGGAGUCAGGUGAGGGGAAGGCGAUUCCCAUCCUGGAGACACCGCAUUAGAGAUGGGGCUGAGUGCCCCUGAGCCUCCAGAUCCCAGGCUAGUGGCGUCUCAUUGGCCUGUCCCACUUGUGUCUGUCUUGGAGCUUUCCUAAUCCCGGGUUUGGCCAGACUCGGCAGGAAUUUCUGUCUGAUUUCAUUAGGGCCCCAAGCCAUUCAUUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
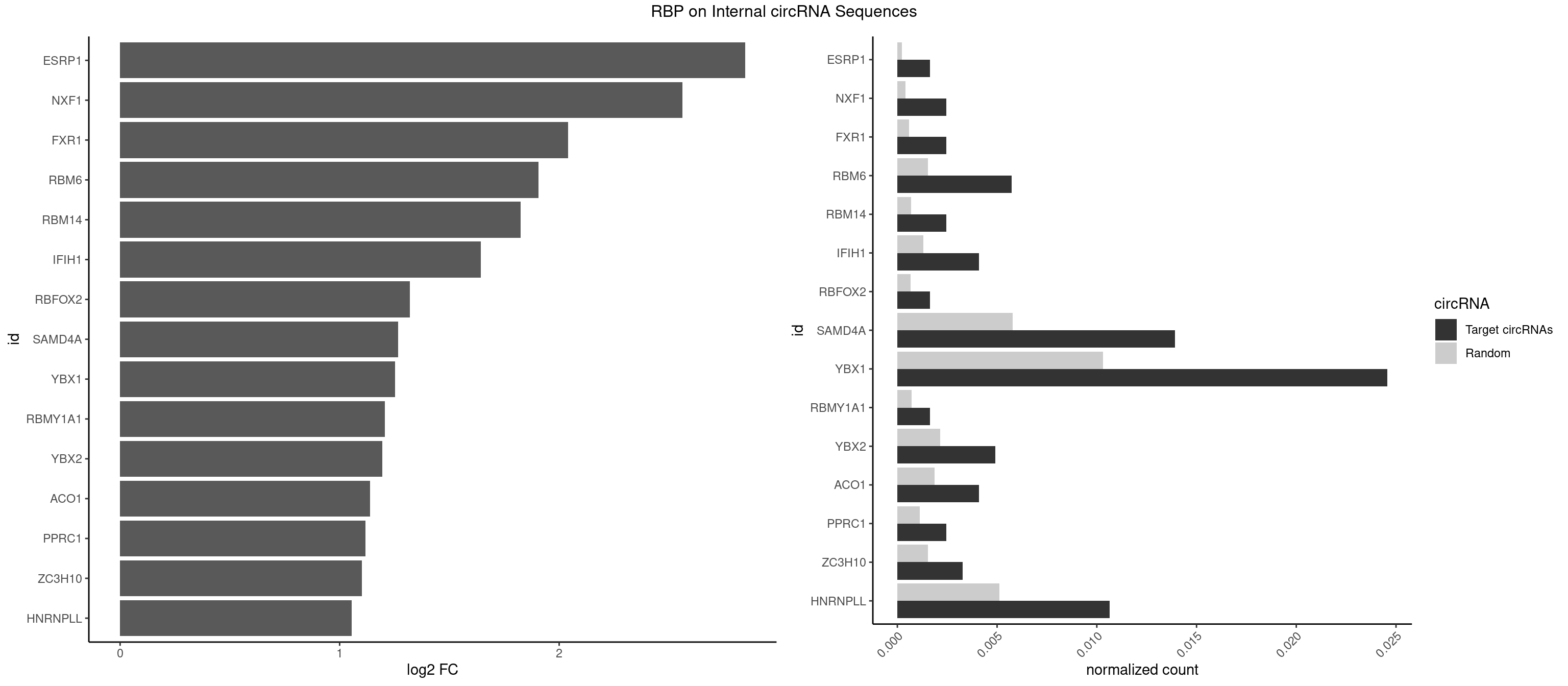
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.001638002 | 0.0002275250 | 2.847840 | AGGGAU | AGGGAU |
| NXF1 | 2 | 286 | 0.002457002 | 0.0004159215 | 2.562516 | AACCUG | AACCUG |
| FXR1 | 2 | 411 | 0.002457002 | 0.0005970720 | 2.040922 | ACGACG,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM6 | 6 | 1054 | 0.005733006 | 0.0015289102 | 1.906788 | AUCCAA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM14 | 2 | 478 | 0.002457002 | 0.0006941687 | 1.823541 | CGCGCC,CGCGGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| IFIH1 | 4 | 904 | 0.004095004 | 0.0013115296 | 1.642615 | GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBFOX2 | 1 | 452 | 0.001638002 | 0.0006564894 | 1.319093 | UGACUG | UGACUG,UGCAUG |
| SAMD4A | 16 | 3992 | 0.013923014 | 0.0057866714 | 1.266666 | CGGGAA,CGGGCC,CGGGUC,CUGGAC,CUGGCA,CUGGCC,CUGGUC,GCGGGA,GCUGGA,GCUGGC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| YBX1 | 29 | 7119 | 0.024570025 | 0.0103183321 | 1.251690 | AACAUC,ACACCA,ACAUCA,ACAUCU,ACCACC,CAACCA,CACACC,CACCAC,CAUCAU,CAUCUG,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GCCUGC,GGUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| RBMY1A1 | 1 | 489 | 0.001638002 | 0.0007101099 | 1.205822 | ACAAGA | ACAAGA,CAAGAC |
| YBX2 | 5 | 1480 | 0.004914005 | 0.0021462711 | 1.195067 | AACAUC,ACAACG,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ACO1 | 4 | 1283 | 0.004095004 | 0.0018607779 | 1.137959 | CAGUGA,CAGUGC,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PPRC1 | 2 | 780 | 0.002457002 | 0.0011318283 | 1.118244 | CCGCGC,CGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| ZC3H10 | 3 | 1053 | 0.003276003 | 0.0015274610 | 1.100801 | CCAGCG,GAGCGA,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| HNRNPLL | 12 | 3534 | 0.010647011 | 0.0051229360 | 1.055406 | ACAAAC,ACACCA,CACACC,CACCAC,CACCGC,CAGACG,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
RBP on BSJ (Exon Only)
Plot
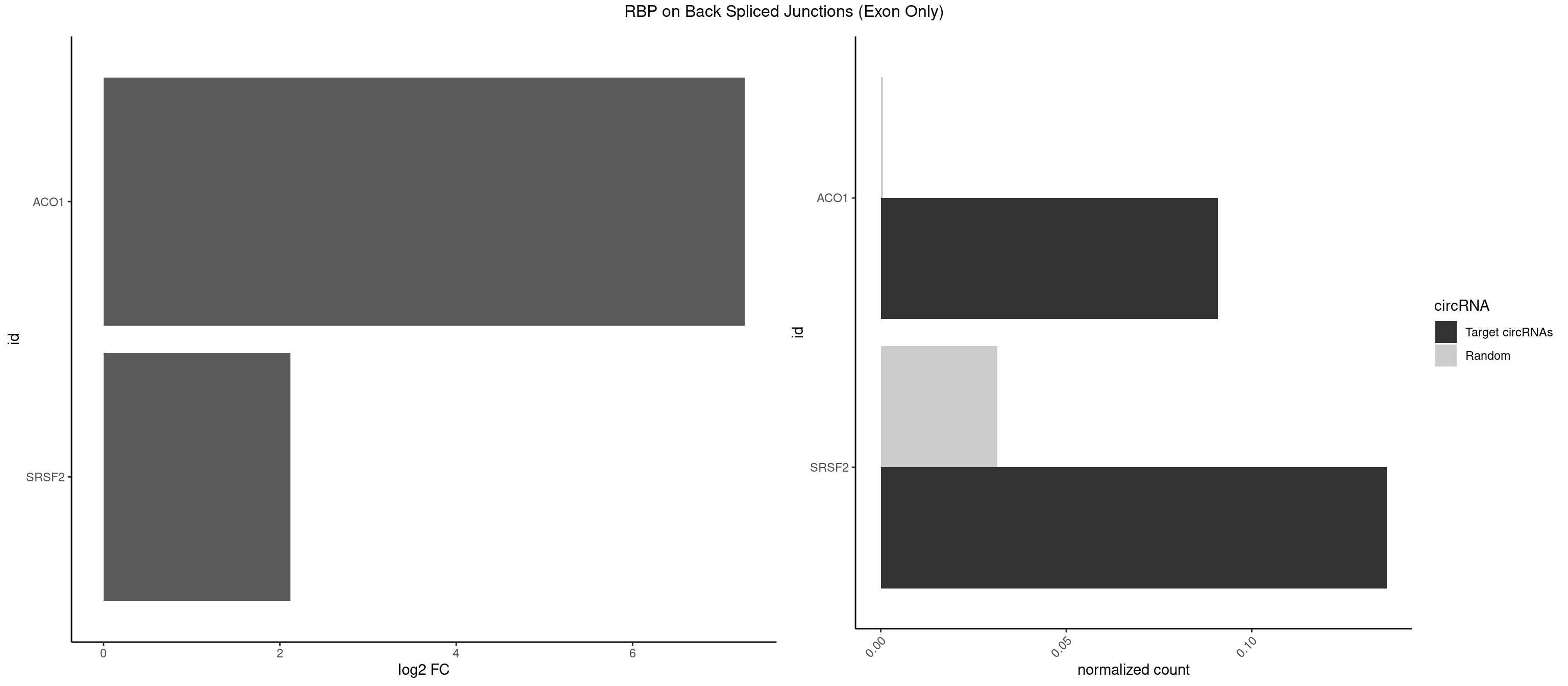
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ACO1 | 1 | 8 | 0.09090909 | 0.0005894682 | 7.268867 | CAGUGC | CAGUGA,CAGUGC,CAGUGG |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | GUCAGU,UCAGUG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
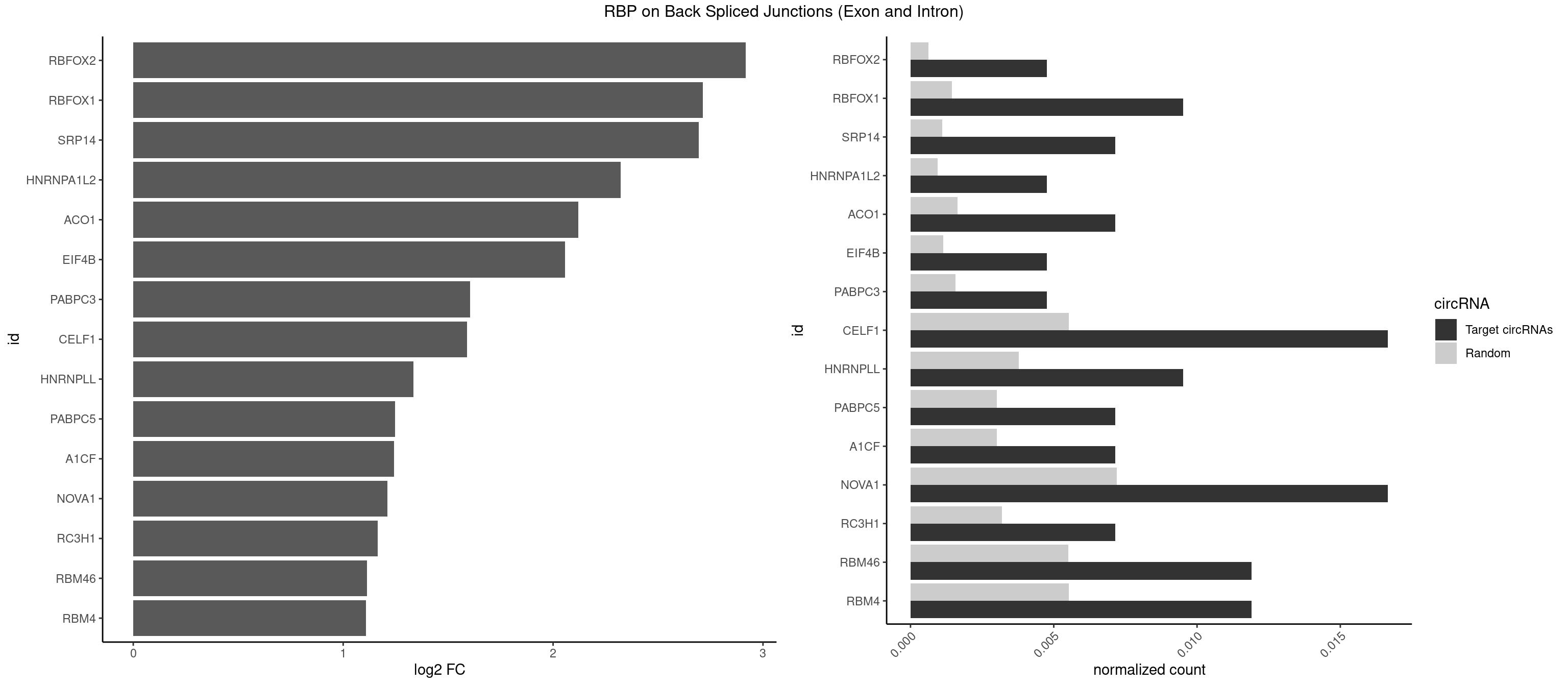
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGA,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | CUGUCU,UGUCUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | ACCGCA,CACCGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | AUUCAU,CAUUCA,UUCAUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AAUGAA,AUCAAU,AUGAAA,AUGAUG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CUCUUU,CUUCCU,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.