circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000124571:-:6:43560177:43561007
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124571:-:6:43560177:43561007 | ENSG00000124571 | ENST00000265351 | - | 6 | 43560178 | 43561007 | 210 | GGUGCAGAUUCUGAUGUAGAAACACCAUCAAACUUUGGAAAAUACCUGGAAUCUUUUCUUGCUUUCACAACCCAUCCAAGUCAGUUUCUACGCUCUUCAACUCAGAUGACUUGGGGAGCCCUCUUCAGGCAUGAAAUCCUGUCCCGUGAUCCUUUGCUAUUAGCAAUAAUACCAAAAUAUCUUCGUGCUUCCAUGACUAACUUGGUCAAGGGUGCAGAUUCUGAUGUAGAAACACCAUCAAACUUUGGAAAAUACCUGGA | circ |
| ENSG00000124571:-:6:43560177:43561007 | ENSG00000124571 | ENST00000265351 | - | 6 | 43560178 | 43561007 | 22 | ACUUGGUCAAGGGUGCAGAUUC | bsj |
| ENSG00000124571:-:6:43560177:43561007 | ENSG00000124571 | ENST00000265351 | - | 6 | 43560998 | 43561207 | 210 | UAAUGAUGAUGAUUGAGUAGGCAAAGUAACCUAUAUUUUAGCCCUUGAAGUGAUGCCUUUCUUUAGCCAAAAGGAAAUACAAAGUCCUUUUGAAACAACAUUUUUUGAGGGUUUAAUUUAUCUGUUUUUAUUACUUCCCUGCUUUUUCCUCUCCUGCUUUUCUCGAUCAACACCGUUAAUAUAGUGGUCUUCUCCUACAGGGUGCAGAUU | ie_up |
| ENSG00000124571:-:6:43560177:43561007 | ENSG00000124571 | ENST00000265351 | - | 6 | 43559978 | 43560187 | 210 | CUUGGUCAAGGUAUAUAAUGGGAAUCUAAACAUGUGGAAUGUAGGUGAAUACACAUAAUAAGACUUUGAGGCUGGGUGCGGUGGCUCACGCCUAUAAUCCCAGCUCUAUGGAAGGCUGAGGCGGGAGGAUCGCUUGAGCCCAGGAGUUGGAGACCAGCCUGUGCGACAUGGCAAAACCUCGUCUCUACUAAAAAUACAAAAAAAUUAGCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
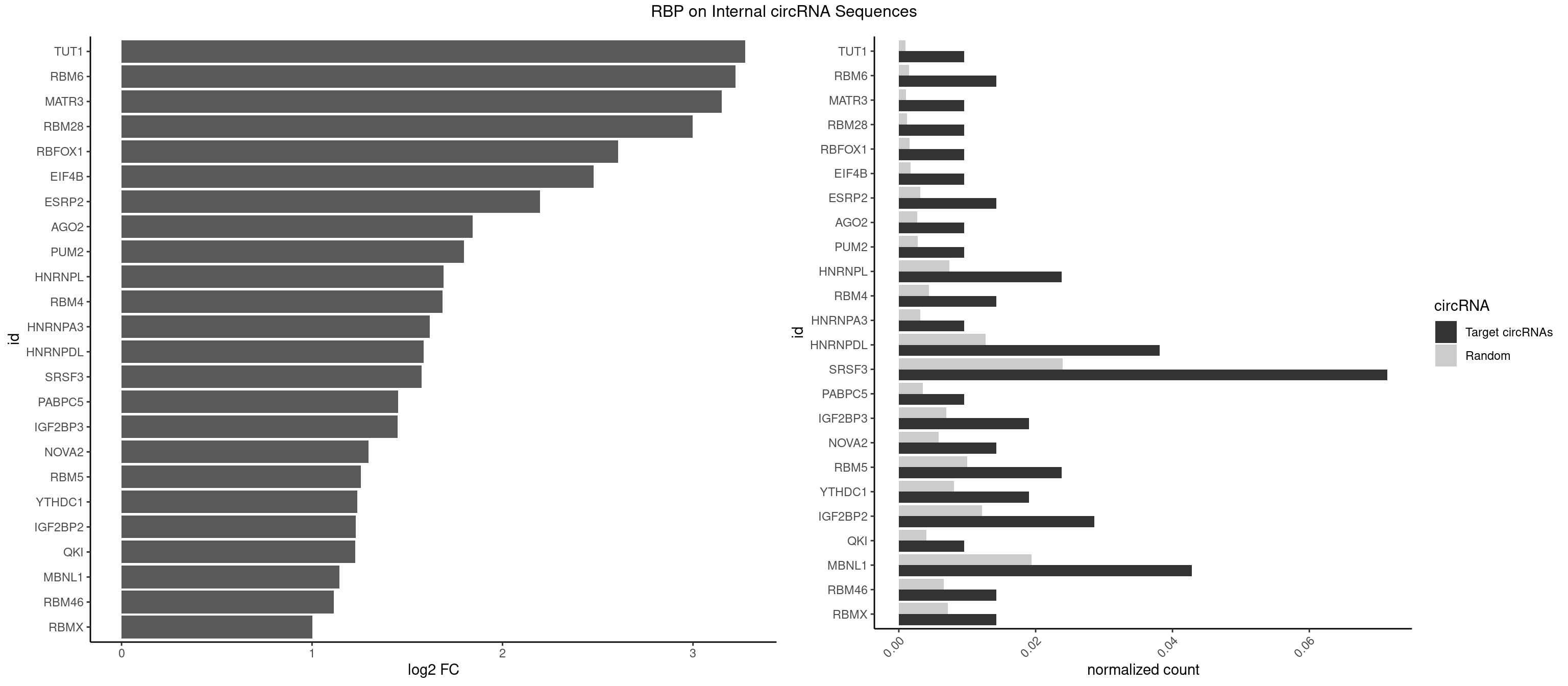
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TUT1 | 1 | 678 | 0.00952381 | 0.0009840095 | 3.274795 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.01428571 | 0.0015289102 | 3.223998 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 1 | 739 | 0.00952381 | 0.0010724109 | 3.150681 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM28 | 1 | 822 | 0.00952381 | 0.0011926949 | 2.997314 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBFOX1 | 1 | 1077 | 0.00952381 | 0.0015622419 | 2.607921 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| EIF4B | 1 | 1179 | 0.00952381 | 0.0017100607 | 2.477491 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ESRP2 | 2 | 2150 | 0.01428571 | 0.0031172377 | 2.196233 | GGGGAG,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| AGO2 | 1 | 1830 | 0.00952381 | 0.0026534924 | 1.843646 | GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PUM2 | 1 | 1890 | 0.00952381 | 0.0027404447 | 1.797129 | UGUAGA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPL | 4 | 5085 | 0.02380952 | 0.0073706513 | 1.691675 | AAACAC,AAAUAC,ACACCA,CACAAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBM4 | 2 | 3065 | 0.01428571 | 0.0044432593 | 1.684883 | CCUCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPA3 | 1 | 2140 | 0.00952381 | 0.0031027457 | 1.617993 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPDL | 7 | 8759 | 0.03809524 | 0.0126950266 | 1.585347 | AAUACC,ACACCA,ACUAAC,AUACCA,AUUAGC,CUAACU | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| SRSF3 | 14 | 16536 | 0.07142857 | 0.0239654858 | 1.575543 | AACUUU,AUCUUC,CACAAC,CAUCAA,CCUCUU,CUCUUC,CUUCAA,CUUCAG,UCACAA,UCUUCA,UCUUCG,UUCAAC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| PABPC5 | 1 | 2400 | 0.00952381 | 0.0034795387 | 1.452643 | GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| IGF2BP3 | 3 | 4815 | 0.01904762 | 0.0069793662 | 1.448443 | AAACAC,AAAUAC,AACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| NOVA2 | 2 | 4013 | 0.01428571 | 0.0058171047 | 1.296200 | AACACC,AGGCAU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RBM5 | 4 | 6879 | 0.02380952 | 0.0099705232 | 1.255798 | AGGGUG,CAAGGG,CUCUUC | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| YTHDC1 | 3 | 5576 | 0.01904762 | 0.0080822104 | 1.236789 | GGGUGC,UAAUAC,UCGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| IGF2BP2 | 5 | 8408 | 0.02857143 | 0.0121863560 | 1.229306 | AAACAC,AAAUAC,AACUCA,CAACUC,GAAAUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| QKI | 1 | 2805 | 0.00952381 | 0.0040664663 | 1.227763 | ACUAAC | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| MBNL1 | 8 | 13372 | 0.04285714 | 0.0193802045 | 1.144952 | ACGCUC,CUUGCU,GUGCUU,UGCUUC,UGCUUU,UUGCUA,UUGCUU,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| RBM46 | 2 | 4554 | 0.01428571 | 0.0066011240 | 1.113790 | AUCAAA,AUGAAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMX | 2 | 4925 | 0.01428571 | 0.0071387787 | 1.000824 | ACCAAA,AUCAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
RBP on BSJ (Exon Only)
Plot
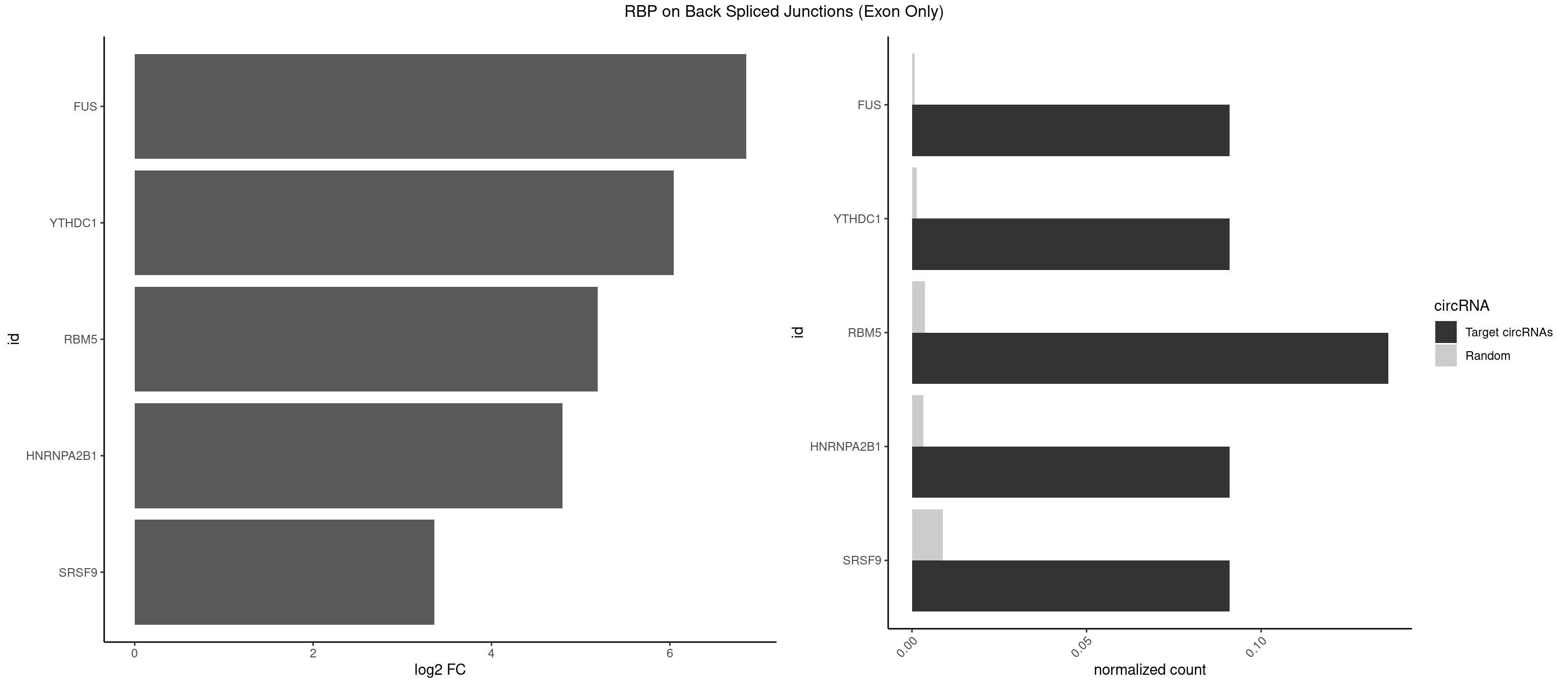
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FUS | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | GGGUGC | AAAAAA,CGGUGG,GGGUGA,GGGUGG,GGGUGU |
| YTHDC1 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | GGGUGC | GAAUAC,GAGUAC,GCCUGC,GGCUGC,GGGUAC,UAAUAC,UAAUGC,UCAUAC,UCCUGC,UCGUGC,UGAUAC,UGCUGC,UGGUGC |
| RBM5 | 2 | 56 | 0.13636364 | 0.0037332984 | 5.190864 | AGGGUG,CAAGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | CAAGGG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| SRSF9 | 1 | 134 | 0.09090909 | 0.0088420225 | 3.361976 | GGGUGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
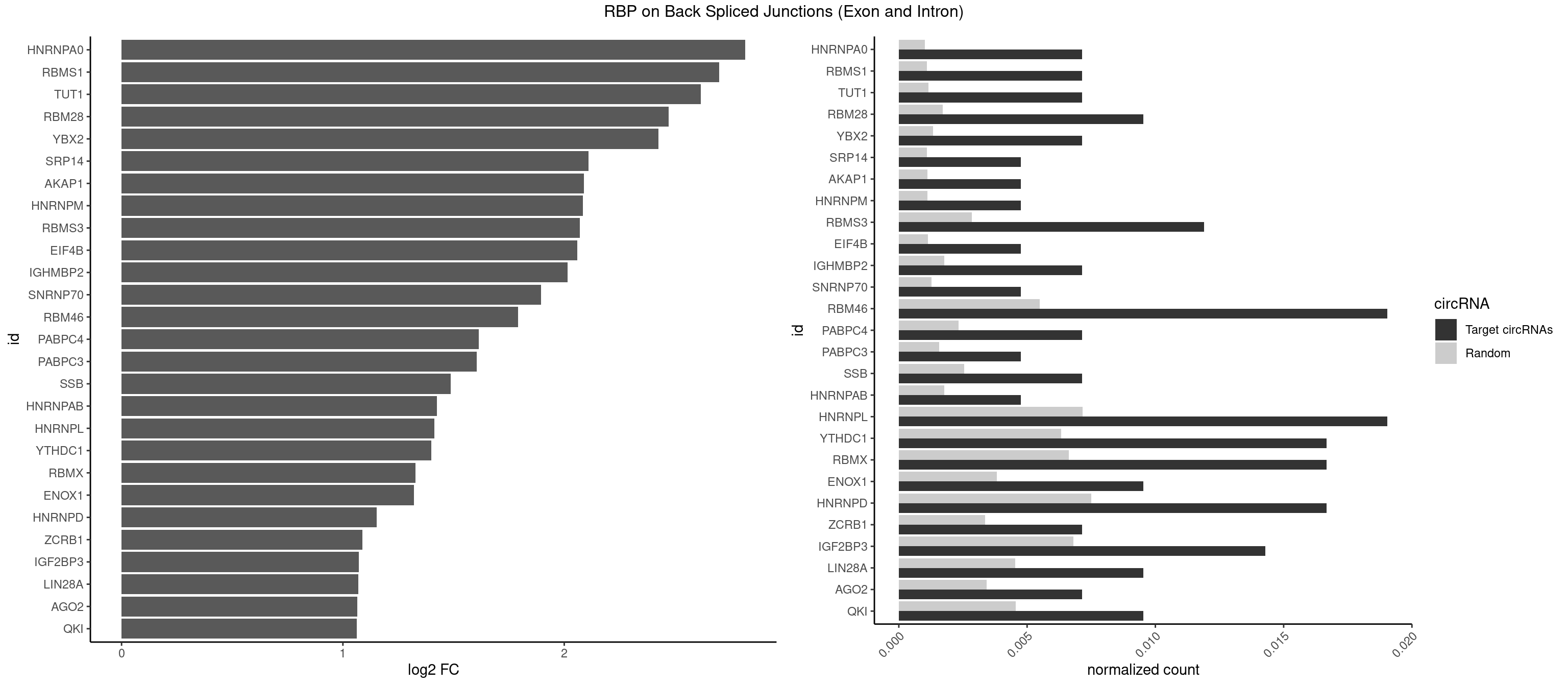
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.001012696 | 2.818299 | AAUUUA,AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.001098347 | 2.701167 | AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| TUT1 | 2 | 230 | 0.007142857 | 0.001163845 | 2.617602 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM28 | 3 | 340 | 0.009523810 | 0.001718057 | 2.470761 | AGUAGG,GAGUAG,UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| YBX2 | 2 | 263 | 0.007142857 | 0.001330109 | 2.424957 | AACAAC,ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| SRP14 | 1 | 218 | 0.004761905 | 0.001103386 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.001118501 | 2.089973 | UAUAUA | AUAUAU,UAUAUA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.001123539 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| RBMS3 | 4 | 562 | 0.011904762 | 0.002836558 | 2.069326 | AAUAUA,AUAUAG,CUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| EIF4B | 1 | 226 | 0.004761905 | 0.001143692 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| IGHMBP2 | 2 | 350 | 0.007142857 | 0.001768440 | 2.014024 | AAAAAA | AAAAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM46 | 7 | 1091 | 0.019047619 | 0.005501814 | 1.791631 | AAUGAU,AUGAUG,AUGAUU,GAUCAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.002332729 | 1.614483 | AAAAAA | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.001566909 | 1.603618 | AAAACC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SSB | 2 | 505 | 0.007142857 | 0.002549375 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.001773478 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPL | 7 | 1419 | 0.019047619 | 0.007154373 | 1.412713 | AAACAA,AAAUAC,AAUACA,ACAUAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.006323055 | 1.398272 | GAAUAC,GGGUGC,UCCUAC,UCCUGC,UGAUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBMX | 6 | 1316 | 0.016666667 | 0.006635429 | 1.328704 | AAGGAA,AAGUAA,AGUAAC,AUCCCA,GGAAGG,UAAGAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.003813986 | 1.320239 | AAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.007502015 | 1.151615 | AAAAAA,AAUUUA,AGUAGG,AUUUAU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.006796655 | 1.071676 | AAAUAC,AAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| LIN28A | 3 | 900 | 0.009523810 | 0.004539500 | 1.069005 | AGGAGU,GGAGGA,UGGAGA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| AGO2 | 2 | 677 | 0.007142857 | 0.003415961 | 1.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| QKI | 3 | 904 | 0.009523810 | 0.004559653 | 1.062615 | AUCUAA,UAACCU,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.