circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000061938:-:3:195872275:195888606
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000061938:-:3:195872275:195888606 | ENSG00000061938 | ENST00000333602 | - | 3 | 195872276 | 195888606 | 1469 | AGGCUGGGAGGCGGCAGAAUGCAGCCAGAGGAGGGCACAGGCUGGCUGCUGGAGCUGCUGUCCGAGGUGCAGCUGCAACAGUACUUCCUGCGGCUCCGAGAUGACCUCAACGUCACCCGCCUGUCCCACUUUGAGUACGUCAAGAAUGAGGACCUGGAGAAGAUCGGCAUGGGUCGGCCUGGCCAGCGGCGGCUGUGGGAGGCUGUGAAGAGGAGGAAGGCCUUGUGCAAACGCAAGUCGUGGAUGAGUAAGGUGUUCAGUGGAAAGCGACUGGAGGCUGAGUUCCCACCUCAUCACUCUCAGAGCACCUUCCGGAAGACCUCGCCCGCCCCUGGGGGCCCAGCAGGGGAGGGGCCCCUGCAGAGCCUCACCUGCCUCAUUGGGGAGAAGGACCUGCGCCUCCUGGAGAAGCUGGGUGAUGGUUCCUUUGGCGUGGUGCGCAGGGGCGAGUGGGACGCGCCCUCAGGGAAGACGGUGAGUGUGGCUGUGAAGUGCCUGAAGCCCGAUGUCCUGAGCCAGCCAGAAGCCAUGGACGACUUCAUCCGGGAGGUCAAUGCCAUGCACUCGCUCGACCACCGAAACCUCAUCCGCCUCUACGGGGUGGUGCUCACGCCGCCCAUGAAGAUGGUGACAGAGCUGGCACCUCUGGGAUCGUUGUUGGACCGGCUACGUAAGCACCAGGGCCACUUCCUCCUGGGGACUCUGAGCCGCUACGCUGUGCAGGUGGCUGAGGGCAUGGGCUACCUGGAGUCCAAGCGCUUUAUUCACCGUGACCUGGCUGCCCGCAAUCUGCUGUUGGCUACCCGCGACCUGGUCAAGAUCGGGGACUUUGGGCUGAUGCGAGCACUACCUCAGAAUGACGACCAUUACGUCAUGCAGGAACAUCGCAAGGUGCCCUUCGCCUGGUGUGCCCCCGAGAGCCUGAAGACACGCACCUUCUCCCAUGCCAGCGACACCUGGAUGUUCGGGGUGACACUGUGGGAAAUGUUCACCUACGGCCAGGAGCCCUGGAUCGGCCUCAACGGCAGUCAGAUCCUGCAUAAGAUCGACAAGGAGGGGGAGCGGCUGCCCCGGCCCGAGGACUGUCCCCAGGACAUCUACAACGUCAUGGUCCAGUGCUGGGCUCACAAGCCAGAGGACAGACCCACGUUUGUGGCCCUGCGGGACUUCCUGCUGGAGGCCCAGCCCACAGACAUGCGGGCCCUUCAGGACUUUGAGGAACCGGACAAGCUGCACAUCCAGAUGAAUGAUGUCAUCACCGUCAUCGAGGGAAGGGCCGAGAACUACUGGUGGCGUGGCCAGAACACACGGACGCUGUGUGUGGGGCCCUUCCCUCGCAACGUGGUGACCUCCGUGGCCGGCCUGUCGGCCCAGGACAUCAGCCAGCCCCUGCAGAACAGCUUCAUCCACACAGGGCAUGGCGACAGUGACCCCCGCCACUGCUGGGGCUUCCCGGACAGGAUUGACGAAGGCUGGGAGGCGGCAGAAUGCAGCCAGAGGAGGGCACAGGCUGGCUGCU | circ |
| ENSG00000061938:-:3:195872275:195888606 | ENSG00000061938 | ENST00000333602 | - | 3 | 195872276 | 195888606 | 22 | AGGAUUGACGAAGGCUGGGAGG | bsj |
| ENSG00000061938:-:3:195872275:195888606 | ENSG00000061938 | ENST00000333602 | - | 3 | 195888597 | 195888806 | 210 | CGCCCUAAUUGUGCCCUGUGGCUGGCAGGAAUAGGGGACGUGGGUCCAGGCGGUUCUCGGUAGCUUCCCUCGGGAGUCAGAAGGUGGUGUGAGCCCUUCCGGGUGGCCGUGUAGUGGGAUGAAGGUGAGCCCAGGUCACUCGGGCAGGCCCCUGUUGUCCCCCCACAACCCUUGUCCCUGAGCCACUCCUCCCCCCACAGAGGCUGGGAG | ie_up |
| ENSG00000061938:-:3:195872275:195888606 | ENSG00000061938 | ENST00000333602 | - | 3 | 195872076 | 195872285 | 210 | GGAUUGACGAGUGAGUACUGGGCGGGGAUGGAUGCUGACCUGGCCCCGCUCGGGACUCCAGGGCCCUCAGCACGGCACGCGUGGGCGGAAUCUGCUCUUCACCCUUUCGCCCGGGCUUCAGCCCCCGACCCGCCACUCUCCCCUCCUCACCCUGGCCUUACUCUCGGGAAGCUGUGGAACCCUAGGAAGCCCAGCCUGGUGGGGGUUCUC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
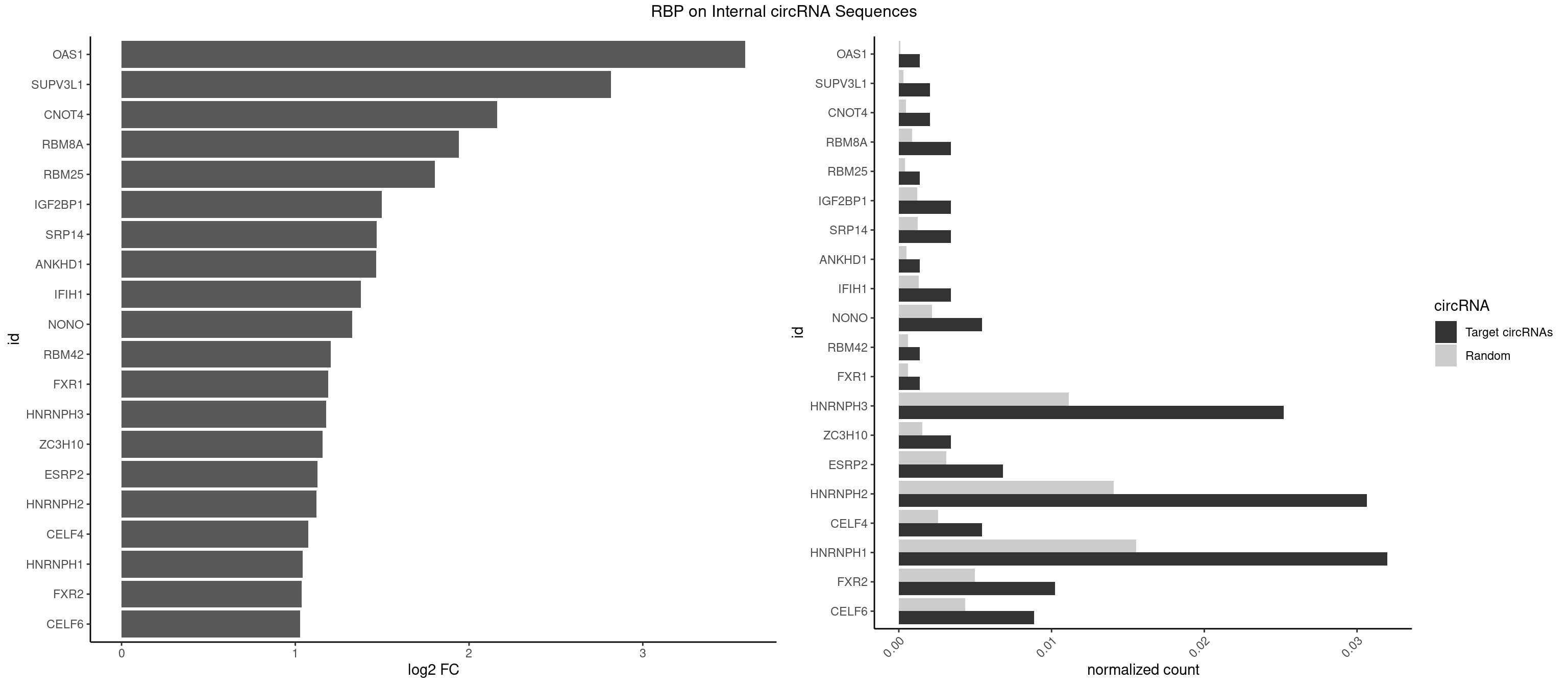
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.001361470 | 0.0001130379 | 3.590287 | GCAUAA | GCAUAA |
| SUPV3L1 | 2 | 199 | 0.002042206 | 0.0002898408 | 2.816796 | CCGCCC | CCGCCC |
| CNOT4 | 2 | 314 | 0.002042206 | 0.0004564992 | 2.161444 | GACAGA | GACAGA |
| RBM8A | 4 | 611 | 0.003403676 | 0.0008869128 | 1.940229 | ACGCGC,CGCGCC,GUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM25 | 1 | 268 | 0.001361470 | 0.0003898359 | 1.804227 | AUCGGG | AUCGGG,CGGGCA,UCGGGC |
| IGF2BP1 | 4 | 831 | 0.003403676 | 0.0012057377 | 1.497178 | AAGCAC,AGCACC,CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRP14 | 4 | 847 | 0.003403676 | 0.0012289250 | 1.469697 | CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ANKHD1 | 1 | 339 | 0.001361470 | 0.0004927293 | 1.466298 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| IFIH1 | 4 | 904 | 0.003403676 | 0.0013115296 | 1.375843 | GGCCCU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| NONO | 7 | 1498 | 0.005445882 | 0.0021723567 | 1.325905 | AGAGGA,AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM42 | 1 | 407 | 0.001361470 | 0.0005912752 | 1.203264 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.001361470 | 0.0005970720 | 1.189189 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPH3 | 36 | 7688 | 0.025187202 | 0.0111429292 | 1.176562 | AAGGUG,AGGGAA,AGGGGA,AUCGGG,AUUGGG,CGAGGG,GAAGGG,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUCG,UCGAGG,UGGGUG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| ZC3H10 | 4 | 1053 | 0.003403676 | 0.0015274610 | 1.155958 | CAGCGA,CCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| ESRP2 | 9 | 2150 | 0.006807352 | 0.0031172377 | 1.126826 | GGGAAA,GGGAAG,GGGGAG,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPH2 | 44 | 9711 | 0.030633084 | 0.0140746688 | 1.121990 | AAGGUG,AGGGAA,AGGGGA,AUCGGG,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUCG,UCGAGG,UGGGGG,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| CELF4 | 7 | 1782 | 0.005445882 | 0.0025839306 | 1.075598 | GGUGUG,GGUGUU,GUGUGG,GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPH1 | 46 | 10717 | 0.031994554 | 0.0155325680 | 1.042530 | AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUCGGG,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAGGAA,GAGGGG,GGAAGG,GGAGGA,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUCG,UCGAGG,UGGGGG,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| FXR2 | 14 | 3434 | 0.010211028 | 0.0049780156 | 1.036485 | AGACGG,GACAAG,GACAGA,GACAGG,GACGAA,GGACAA,GGACAG,GGACGA,UGACAG,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CELF6 | 12 | 2997 | 0.008849558 | 0.0043447134 | 1.026344 | GUGAUG,GUGGGG,GUGGUG,GUGUGG,UGUGGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
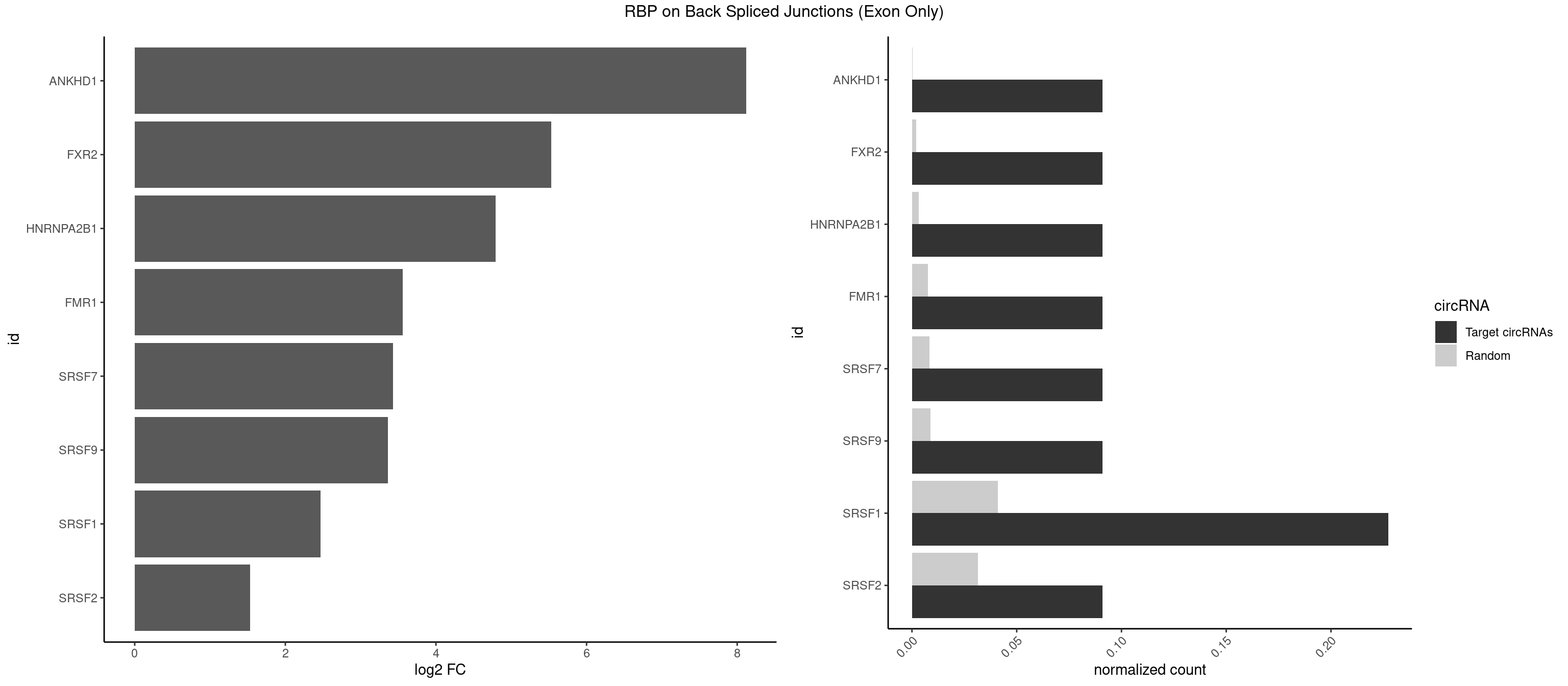
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 4 | 0.09090909 | 0.0003274823 | 8.116864 | GACGAA | AGACGU,GACGAA,GACGUA,GACGUU |
| FXR2 | 1 | 29 | 0.09090909 | 0.0019648939 | 5.531901 | GACGAA | AGACAA,AGACAG,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGGA,GGACAA,GGACAG,GGACGA,UGACAA,UGACAG,UGACGA |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | CGAAGG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| FMR1 | 1 | 117 | 0.09090909 | 0.0077285827 | 3.556149 | CGAAGG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| SRSF7 | 1 | 128 | 0.09090909 | 0.0084490438 | 3.427565 | GAAGGC | AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACUACG,AGAAGA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CGAAUG,CUCUUC,CUGAGA,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UAGAGA,UCUUCA,UGAGAG,UGGACA |
| SRSF9 | 1 | 134 | 0.09090909 | 0.0088420225 | 3.361976 | GAAGGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAC,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCG,GGAGGA,GGAGGC,GGAUGG,GGGAGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGAGAA,UGAGAG,UGAGCA,UGAUGG,UGGAGC,UGGAGG,UGGUGC |
| SRSF1 | 4 | 625 | 0.22727273 | 0.0410007860 | 2.470701 | ACGAAG,CGAAGG,GAAGGC,GACGAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | GAAGGC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
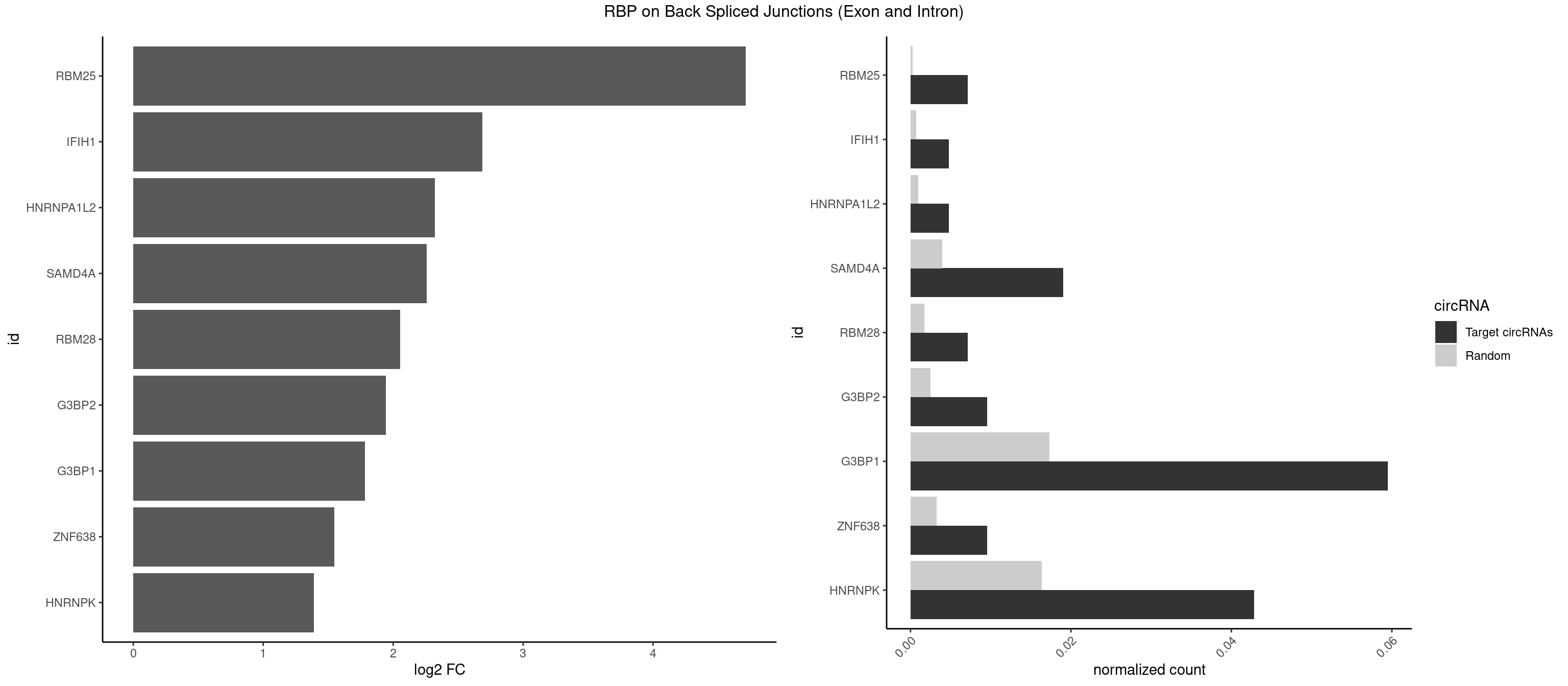
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 2 | 53 | 0.007142857 | 0.0002720677 | 4.714464 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SAMD4A | 7 | 790 | 0.019047619 | 0.0039852882 | 2.256855 | CGGGAA,CGGGAC,CGGGCA,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | GUGUAG,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | GGAUGA,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| G3BP1 | 24 | 3431 | 0.059523810 | 0.0172914148 | 1.783411 | ACCCGC,AGGCCC,CAGGCC,CCACAG,CCAGGC,CCCACA,CCCAGG,CCCCAC,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCUC,CCCGCC,CCCUAG,CCCUCC,CCCUCG,CCUAGG,CCUCGG,CGGCAC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPK | 17 | 3244 | 0.042857143 | 0.0163492543 | 1.390311 | ACCCGC,ACCCUU,CAACCC,CCCCAC,CCCCCA,CCCCCC,GCCCAG,GCCCCC,GCCCCU,UCCCCC,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.