circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000009307:-:1:114719578:114725333
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000009307:-:1:114719578:114725333 | ENSG00000009307 | ENST00000690784 | - | 1 | 114719579 | 114725333 | 1372 | UGAGUUCUCUGGUGAUGUUGAUAGCCUGGAACUGGGGGACAUGGUCGAGUAUAGCUUGUCCAAAGGCAAAGGCAACAAAGUCAGUGCAGAAAAAGUGAACAAAACACACUCAGUGAAUGGCAUUACUGAGGAAGCUGAUCCCACCAUUUACUCUGGCAAAGUAAUUCGCCCCCUGAGGAGUGUUGAUCCAACACAGACUGAGUACCAAGGAAUGAUUGAGAUUGUGGAGGAGGGCGAUAUGAAAGGUGAGGUCUAUCCAUUUGGCAUCGUUGGGAUGGCCAACAAAGGGGAUUGCCUGCAGAAAGGGGAGAGCGUCAAGUUCCAAUUGUGUGUCCUGGGCCAAAAUGCACAAACUAUGGCUUACAACAUCACACCCCUGCGCAGGGCCACAGUGGAAUGUGUGAAAGAUCAGGUAAGUGCCAGCAUCUCUGUAUCUGAAUUUGAUCCUUCUAUGAGUUGGUAACCAAAACCUUCAAAUAUUUUCAGCCAAGGGGAAAUCAUCAACAUUCAUUCGUUUUUUUAUUUCUACUUCACCUUUUUUCAGUCAUUUGAGGUAGCUUAAUAAAAACAGAAAACUACACAAAAAGUGACUAUAUUAGCAAUGAGAAAGUAAGAUUGGUAUUAGUAACUAAUAUUAUGCAAUGGGGAAAAUAAGUUAGGGGUGAGAUUUCCAGAAUAGAAGGACUAUAUGGUUUUGUACACUUUUGCUUAGAGCUUCUUUUGCAUAUCACACAAACACAGAAACCACGUUCAAAAGUACUUAAAAAAUAAGCUUAGGGCAAACAAACUUAGUCUUACUCCUGAGAUCUCAGGGAAAUCUGAGUCCUCAGAAUGUUUUUCAUCAGUACAGAGAAUAAUAACAUUUUGUAUGGCUGCUACUAUAACCCAGCUUAAUUCUAGAGGGGAUCCAAACAAUAUAUAUGUUGAAGUAACAGCUUCCUGAUGAGUUUGCUUAAUUCAAAAAGCUUAGUUCAUAAUAUACGAAGGAACAAGGCAAGUUUUAUUUGUUUGGUUCAAGUUUGUGAUAUAGUGAAUGGCACUUUCAGAGAUACAUUUCUUCCCUGACAGGUUUUGCCACAGGCAGCUGGACUGGAGUAUCAACAUUAAUACCAUGUUUUAUACAUCCUUUUAGAUAGGGGCAGGCAUUGUGCUUCAUUCAGAGUGAGGUGUGGCAUGCCCUCUCUUUUUUACCUACUUUUUUUUUUCAGUUUGGCUUCAUUAACUAUGAAGUAGGAGAUAGCAAGAAGCUCUUUUUCCAUGUGAAAGAAGUUCAGGAUGGCAUUGAGCUACAGGCAGGAGAUGAGGUGGAGUUCUCAGUGAUUCUUAAUCAGCGCACUGGCAAGUGCAGCGCCUGUAAUGUUUGGCGAGUCUGUGAGUUCUCUGGUGAUGUUGAUAGCCUGGAACUGGGGGACAUGGUCGAGU | circ |
| ENSG00000009307:-:1:114719578:114725333 | ENSG00000009307 | ENST00000690784 | - | 1 | 114719579 | 114725333 | 22 | UGGCGAGUCUGUGAGUUCUCUG | bsj |
| ENSG00000009307:-:1:114719578:114725333 | ENSG00000009307 | ENST00000690784 | - | 1 | 114725324 | 114725533 | 210 | UAUCCUGAAGUUAGGGAUAAUGUUACCUCUUCAUGGGGCAUUCUUGAGGAUUAAAUGAGUCAAUACAUAGAAAUUACUUAAAAUGGUGUCUAAUGUAAAUAAGCAUGCCAUGCCAUGACUGUUACUGUAAAAUAAAGACUGCCUGUUACUGUUGAUGUUUAUGGUAAUGUUUAGUUAAAUGUCAUUUCCUUUCUCCUUAGUGAGUUCUCU | ie_up |
| ENSG00000009307:-:1:114719578:114725333 | ENSG00000009307 | ENST00000690784 | - | 1 | 114719379 | 114719588 | 210 | GGCGAGUCUGGUGAGUUUUGUUGUUGUGGUUUGAUUAGUAACUACCCUGGAGUGUCUCAACUUUAUAGUCUCUGUUUUGUCACAUCACUUUUCUGUGCCUUGUCUUCUGCCACUUAUUAUUUCUUGAUACUUCCUACUUCUAGUUGAAUACAAAAAGAUUGUUUAGUACAUGUUUUUUAAAAUUUUUGCCAAAUUCUUCUUCUUCCAUUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
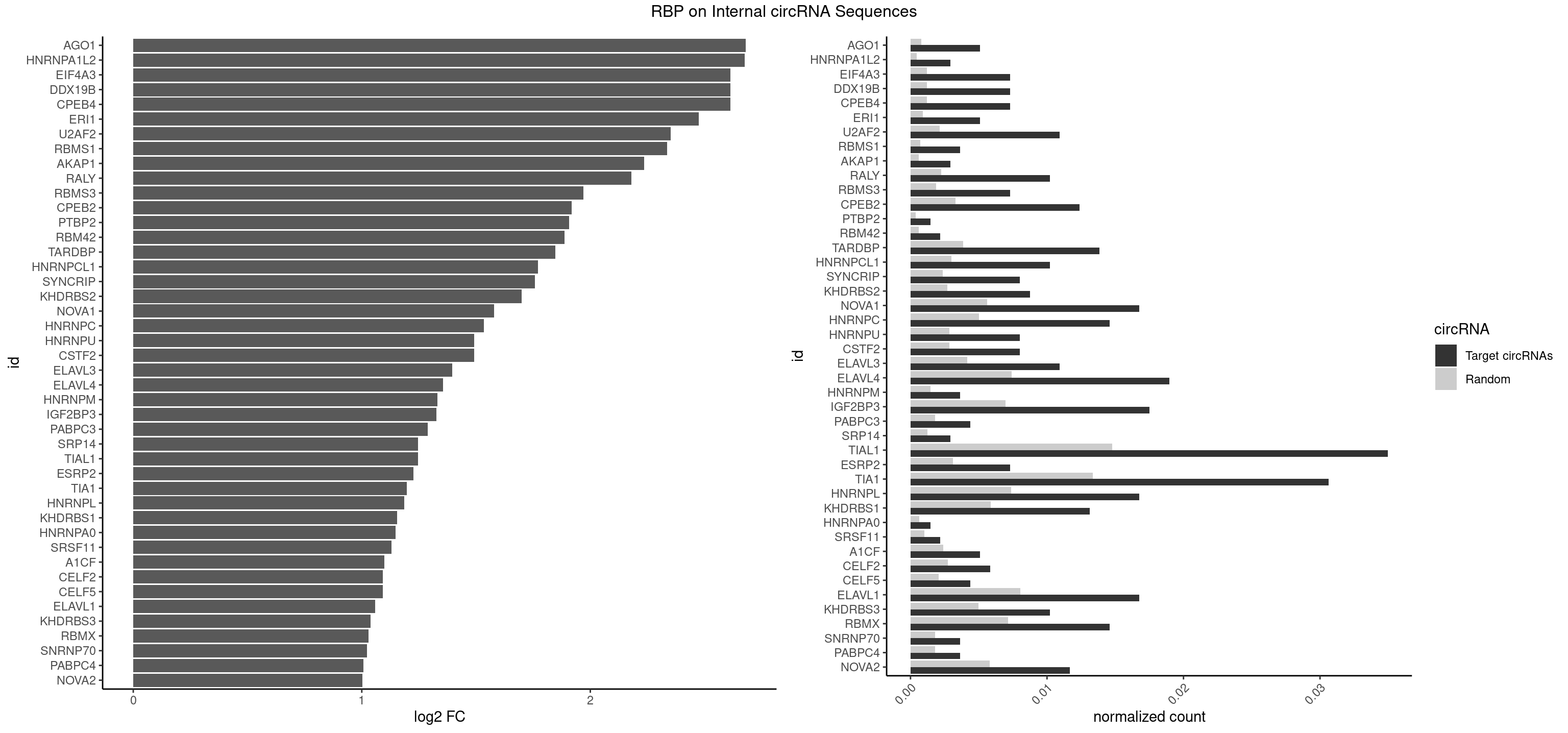
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AGO1 | 6 | 548 | 0.005102041 | 0.0007956130 | 2.680936 | AGGUAG,GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPA1L2 | 3 | 314 | 0.002915452 | 0.0004564992 | 2.675035 | AUAGGG,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| CPEB4 | 9 | 821 | 0.007288630 | 0.0011912456 | 2.613177 | UUUUUU | UUUUUU |
| DDX19B | 9 | 821 | 0.007288630 | 0.0011912456 | 2.613177 | UUUUUU | UUUUUU |
| EIF4A3 | 9 | 821 | 0.007288630 | 0.0011912456 | 2.613177 | UUUUUU | UUUUUU |
| ERI1 | 6 | 632 | 0.005102041 | 0.0009173461 | 2.475536 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| U2AF2 | 14 | 1477 | 0.010932945 | 0.0021419234 | 2.351703 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBMS1 | 4 | 497 | 0.003644315 | 0.0007217036 | 2.336169 | AUAUAC,AUAUAG,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 3 | 426 | 0.002915452 | 0.0006188101 | 2.236151 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RALY | 13 | 1553 | 0.010204082 | 0.0022520629 | 2.179827 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RBMS3 | 9 | 1283 | 0.007288630 | 0.0018607779 | 1.969742 | AAUAUA,AUAUAG,AUAUAU,CUAUAU,UAUAGC,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CPEB2 | 16 | 2261 | 0.012390671 | 0.0032780993 | 1.918323 | CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PTBP2 | 1 | 267 | 0.001457726 | 0.0003883867 | 1.908154 | CUCUCU | CUCUCU |
| RBM42 | 2 | 407 | 0.002186589 | 0.0005912752 | 1.886780 | AACUAA,AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| TARDBP | 18 | 2654 | 0.013848397 | 0.0038476365 | 1.847675 | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPCL1 | 13 | 2062 | 0.010204082 | 0.0029897078 | 1.771070 | CUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SYNCRIP | 10 | 1634 | 0.008017493 | 0.0023694485 | 1.758600 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 11 | 1858 | 0.008746356 | 0.0026940701 | 1.698895 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| NOVA1 | 22 | 3872 | 0.016763848 | 0.0056127669 | 1.578569 | AUCAAC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,UCAGUC,UCAUUC,UUCAUU,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPC | 19 | 3471 | 0.014577259 | 0.0050316361 | 1.534620 | CUUUUU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPU | 10 | 1965 | 0.008017493 | 0.0028491350 | 1.492627 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| CSTF2 | 10 | 1967 | 0.008017493 | 0.0028520334 | 1.491160 | GUGUGU,GUGUUG,GUUUUG,UGUGUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ELAVL3 | 14 | 2867 | 0.010932945 | 0.0041563169 | 1.395304 | UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ELAVL4 | 25 | 5105 | 0.018950437 | 0.0073996354 | 1.356705 | AAAAAA,GUAUCU,UAUUUU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPM | 4 | 999 | 0.003644315 | 0.0014492040 | 1.330387 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| IGF2BP3 | 23 | 4815 | 0.017492711 | 0.0069793662 | 1.325586 | AAAAAC,AAAACA,AAACAC,AAAUCA,AACACA,AAUUCA,ACAAAC,ACACAC,ACACUC,ACAUUC,CAAACA,CACACA,CACUCA,CAUUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| PABPC3 | 5 | 1234 | 0.004373178 | 0.0017897669 | 1.288910 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRP14 | 3 | 847 | 0.002915452 | 0.0012289250 | 1.246323 | CCUGUA,CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| TIAL1 | 47 | 10182 | 0.034985423 | 0.0147572438 | 1.245331 | AGUUUU,AUUUUC,AUUUUG,CAGUUU,CUUUUA,CUUUUG,CUUUUU,GUUUUU,UAUUUU,UCAGUU,UUAUUU,UUCAGU,UUUAUU,UUUCAG,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ESRP2 | 9 | 2150 | 0.007288630 | 0.0031172377 | 1.225379 | GGGAAA,GGGGAA,GGGGAG,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| TIA1 | 41 | 9206 | 0.030612245 | 0.0133428208 | 1.198045 | AUUUUC,AUUUUG,CUUUUA,CUUUUG,CUUUUU,GUUUUA,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UUAUUU,UUUAUU,UUUUAU,UUUUGU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPL | 22 | 5085 | 0.016763848 | 0.0073706513 | 1.185489 | AAACAA,AAACAC,AAAUAA,AACAAA,AACACA,AAUAAA,ACACAA,ACACAC,CACAAA,CACACA,CACACC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| KHDRBS1 | 17 | 4064 | 0.013119534 | 0.0058910141 | 1.155129 | AUAAAA,AUUUAC,CAAAAU,GAAAAC,UAAAAA,UUAAAA,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| HNRNPA0 | 1 | 453 | 0.001457726 | 0.0006579386 | 1.147695 | AGUAGG | AAUUUA,AGAUAU,AGUAGG |
| SRSF11 | 2 | 688 | 0.002186589 | 0.0009985015 | 1.130845 | AAGAAG | AAGAAG |
| A1CF | 6 | 1642 | 0.005102041 | 0.0023810421 | 1.099481 | AGUAUA,AUCAGU,GAUCAG,UCAGUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CELF2 | 7 | 1886 | 0.005830904 | 0.0027346479 | 1.092364 | AUGUGU,GUCUGU,GUGUGU,UAUGUU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| CELF5 | 5 | 1415 | 0.004373178 | 0.0020520728 | 1.091600 | GUGUGG,GUGUGU,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL1 | 22 | 5554 | 0.016763848 | 0.0080503280 | 1.058234 | UAUUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGGUU,UUGUUU,UUUAUU,UUUGGU,UUUGUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| KHDRBS3 | 13 | 3429 | 0.010204082 | 0.0049707696 | 1.037605 | AAAUAA,AAUAAA,AUAAAA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBMX | 19 | 4925 | 0.014577259 | 0.0071387787 | 1.029970 | AAGAAG,AAGGAA,AAGUAA,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCCCA,GAAGGA,GUAACA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SNRNP70 | 4 | 1237 | 0.003644315 | 0.0017941145 | 1.022376 | AUUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC4 | 4 | 1251 | 0.003644315 | 0.0018144033 | 1.006152 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| NOVA2 | 15 | 4013 | 0.011661808 | 0.0058171047 | 1.003418 | AGAUCA,AGGCAU,AGUCAU,AUCAAC,AUCAUC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
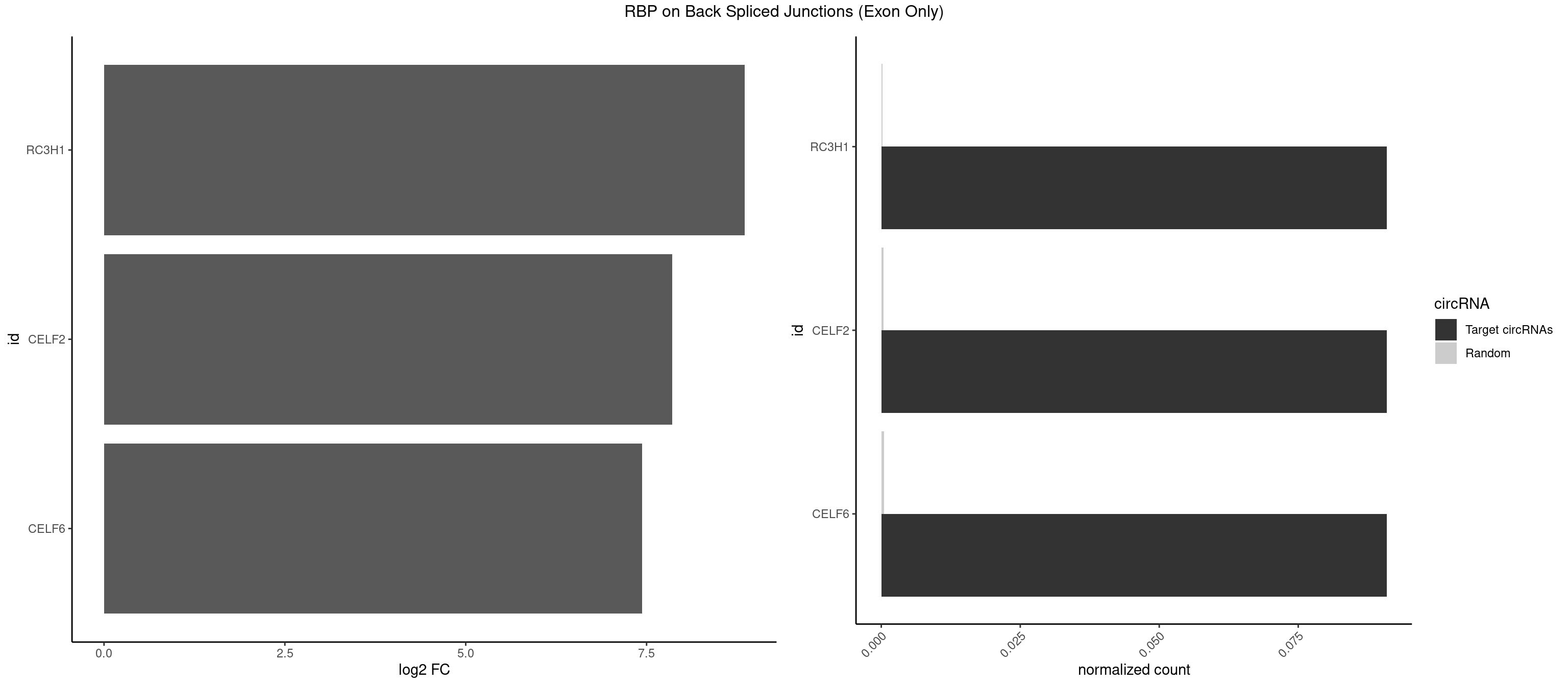
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RC3H1 | 1 | 2 | 0.09090909 | 0.0001964894 | 8.853829 | UCUGUG | CCUUCU,CUUCUG |
| CELF2 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GUCUGU | AUGUGU,GUCUGU,UAUGUU,UGUGUG,UGUUGU |
| CELF6 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | UGUGAG | GUGAGG,GUGUUG,UGUGGG,UGUGUG |
RBP on BSJ (Exon and Intron)
Plot
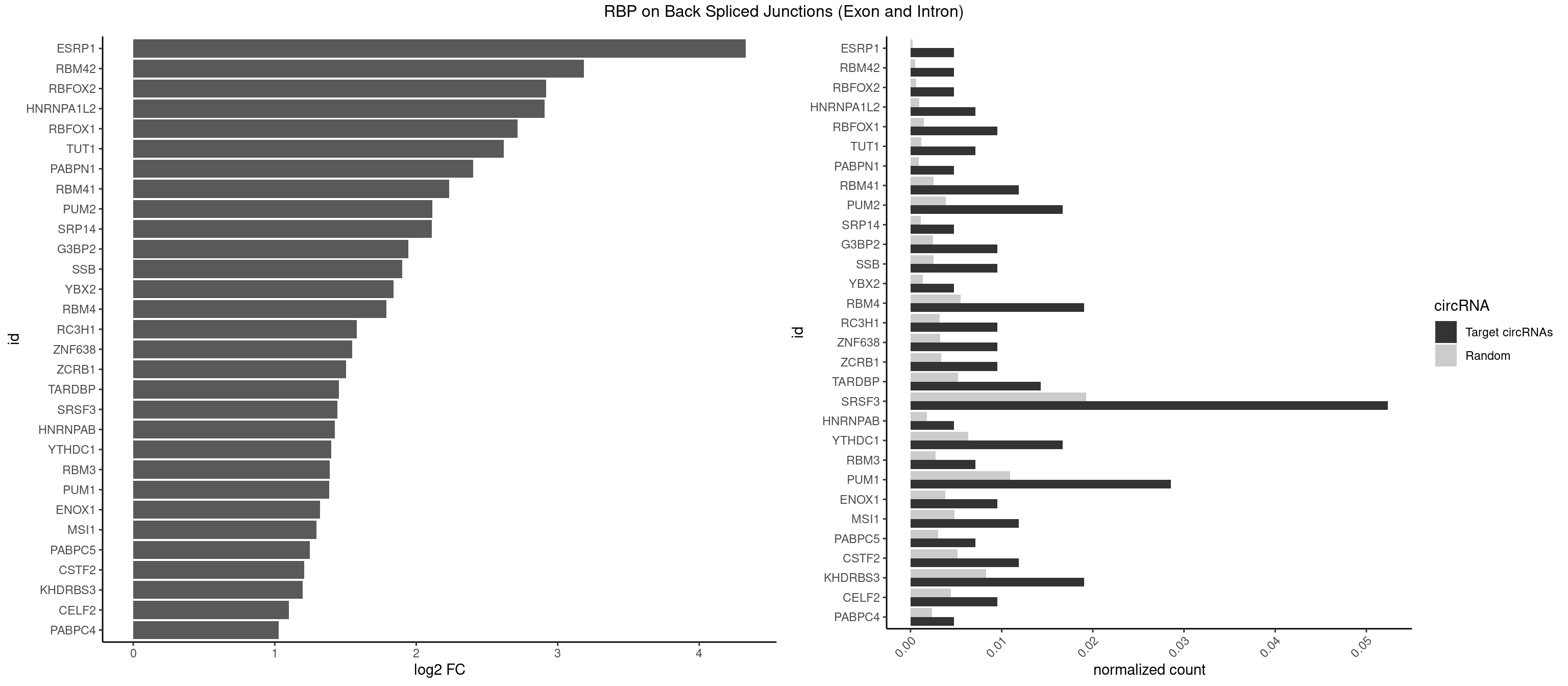
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 46 | 0.004761905 | 0.0002367997 | 4.329800 | AGGGAU | AGGGAU |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGC,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | CAAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACAU,AUACUU,UACAUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PUM2 | 6 | 764 | 0.016666667 | 0.0038542926 | 2.112428 | GUAAAU,GUACAU,UAAAUA,UACAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUAA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM4 | 7 | 1094 | 0.019047619 | 0.0055169287 | 1.787673 | CCUCUU,CUUCCU,CUUCUU,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | ACUUAA,GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GUUGUG,GUUGUU,GUUUUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF3 | 21 | 3825 | 0.052380952 | 0.0192765014 | 1.442199 | AACUUU,ACAUCA,ACUUCU,ACUUUA,CACAUC,CAUCAC,CCUCUU,CUCUUC,CUUCAU,CUUCUU,CUUUAU,UACAAA,UACUUC,UCUUCA,UCUUCC,UUCUCC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.0063230552 | 1.398272 | GAAUAC,GACUGC,GCAUGC,UAGUAC,UCCUAC,UGAUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACU,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AAUGUU,GUAAAU,GUACAU,UAAAUA,UAAUGU,UACAUA,UGUAAA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAUACA,AGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGUUAG,UAGUAA,UAGUUA,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AAAUAA,AAUAAA,AUAAAG,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.