circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000156299:-:21:31135932:31141504
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000156299:-:21:31135932:31141504 | ENSG00000156299 | ENST00000286827 | - | 21 | 31135933 | 31141504 | 408 | CCAAGACAGACACGGCUUUCAAGGCAUUCUUGGAUGCCCAGAACCCGAAGCAGCAGCACUCAUCCACGCUGGAGUCGUACCUCAUCAAGCCCAUCCAGAGGAUCCUCAAGUACCCACUUCUGCUCAGGGAGCUGUUCGCCCUGACCGAUGCGGAGAGCGAGGAGCACUACCACCUGGACGUGGCCAUCAAGACCAUGAACAAGGUUGCCAGUCACAUCAAUGAGAUGCAGAAAAUCCAUGAAGAGUUUGGGGCUGUGUUUGACCAGCUGAUUGCUGAACAGACUGGUGAGAAAAAAGAGGUUGCAGAUCUGAGCAUGGGAGACCUGCUUUUGCACACUACCGUGAUCUGGCUGAACCCGCCGGCCUCGCUGGGCAAGUGGAAAAAGGAACCAGAGUUGGCAGCAUUCGCCAAGACAGACACGGCUUUCAAGGCAUUCUUGGAUGCCCAGAACCCGAAG | circ |
| ENSG00000156299:-:21:31135932:31141504 | ENSG00000156299 | ENST00000286827 | - | 21 | 31135933 | 31141504 | 22 | GGCAGCAUUCGCCAAGACAGAC | bsj |
| ENSG00000156299:-:21:31135932:31141504 | ENSG00000156299 | ENST00000286827 | - | 21 | 31141495 | 31141704 | 210 | CCCCAACACAUAAAUACAUAGAUCUGACUGCCUGUGGUCUCCCUGACUGCCCCCUCCCCUACCUGUCCCUCUUCCUGCCACCCACUCUCUGCUCAGUCCUGCAAAAAGGCACCUUGGAGUCACUGAGGAAGGGAGGGAAAUUGUGGGAAGGGAGUCACGUGGGCGUUCCACCCCCAUGACCUGUGUUUAAACCCUGCCAGCCAAGACAGA | ie_up |
| ENSG00000156299:-:21:31135932:31141504 | ENSG00000156299 | ENST00000286827 | - | 21 | 31135733 | 31135942 | 210 | GCAGCAUUCGGUGUGUAUCAGAAAAGCGUUUUAUGGAGGGGGAAAAGUCUAGCUUAGUUUUCAAAACAAGAACAUUUCUUUUUCAAGACCCAAUUGAUUAAAAAUAAAGUUAAUUGCAGUUACUUGAAUCGGUCUUCCUGCCGUAGCCACCAACAUCUCACAAACACUAAAUGAAACCUUCGAGUUAAAUUGCCCUAUGUGGUCACUCAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
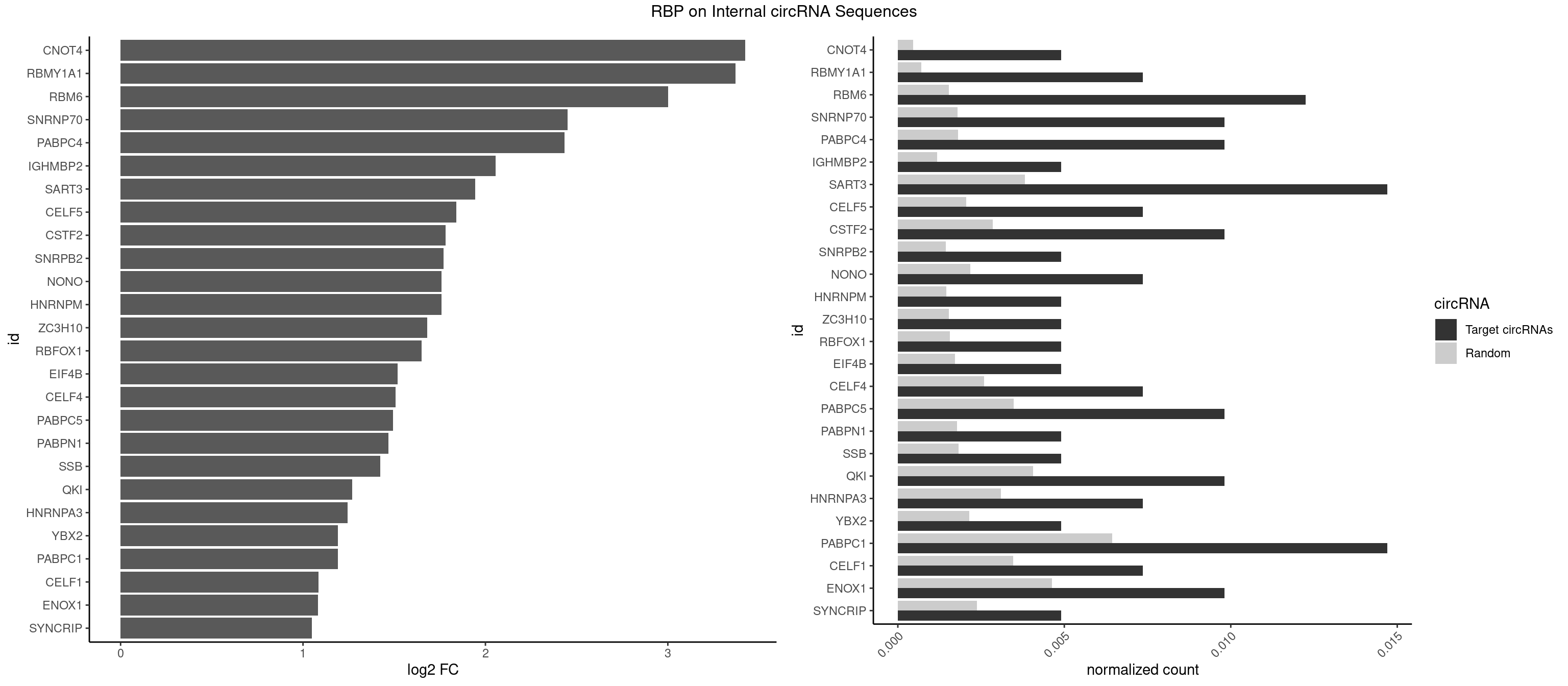
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 314 | 0.004901961 | 0.0004564992 | 3.424675 | GACAGA | GACAGA |
| RBMY1A1 | 2 | 489 | 0.007352941 | 0.0007101099 | 3.372207 | CAAGAC | ACAAGA,CAAGAC |
| RBM6 | 4 | 1054 | 0.012254902 | 0.0015289102 | 3.002783 | AAUCCA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SNRNP70 | 3 | 1237 | 0.009803922 | 0.0017941145 | 2.450087 | AUCAAG,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPC4 | 3 | 1251 | 0.009803922 | 0.0018144033 | 2.433864 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| IGHMBP2 | 1 | 813 | 0.004901961 | 0.0011796520 | 2.054998 | AAAAAA | AAAAAA |
| SART3 | 5 | 2634 | 0.014705882 | 0.0038186524 | 1.945258 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| CELF5 | 2 | 1415 | 0.007352941 | 0.0020520728 | 1.841240 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 3 | 1967 | 0.009803922 | 0.0028520334 | 1.781368 | GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SNRPB2 | 1 | 991 | 0.004901961 | 0.0014376103 | 1.769686 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NONO | 2 | 1498 | 0.007352941 | 0.0021723567 | 1.759060 | AGAGGA,AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPM | 1 | 999 | 0.004901961 | 0.0014492040 | 1.758098 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 1 | 1053 | 0.004901961 | 0.0015274610 | 1.682223 | GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBFOX1 | 1 | 1077 | 0.004901961 | 0.0015622419 | 1.649741 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| EIF4B | 1 | 1179 | 0.004901961 | 0.0017100607 | 1.519311 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF4 | 2 | 1782 | 0.007352941 | 0.0025839306 | 1.508754 | GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC5 | 3 | 2400 | 0.009803922 | 0.0034795387 | 1.494463 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPN1 | 1 | 1222 | 0.004901961 | 0.0017723764 | 1.467674 | AAAAGA | AAAAGA,AGAAGA |
| SSB | 1 | 1260 | 0.004901961 | 0.0018274462 | 1.423530 | GCUGUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| QKI | 3 | 2805 | 0.009803922 | 0.0040664663 | 1.269583 | ACACUA,ACUCAU,CACACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPA3 | 2 | 2140 | 0.007352941 | 0.0031027457 | 1.244776 | AGGAGC,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| YBX2 | 1 | 1480 | 0.004901961 | 0.0021462711 | 1.191527 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PABPC1 | 5 | 4443 | 0.014705882 | 0.0064402624 | 1.191202 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CELF1 | 2 | 2391 | 0.007352941 | 0.0034664959 | 1.084843 | UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ENOX1 | 3 | 3195 | 0.009803922 | 0.0046316558 | 1.081831 | AAGACA,AGACAG,CAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SYNCRIP | 1 | 1634 | 0.004901961 | 0.0023694485 | 1.048808 | AAAAAA | AAAAAA,UUUUUU |
RBP on BSJ (Exon Only)
Plot
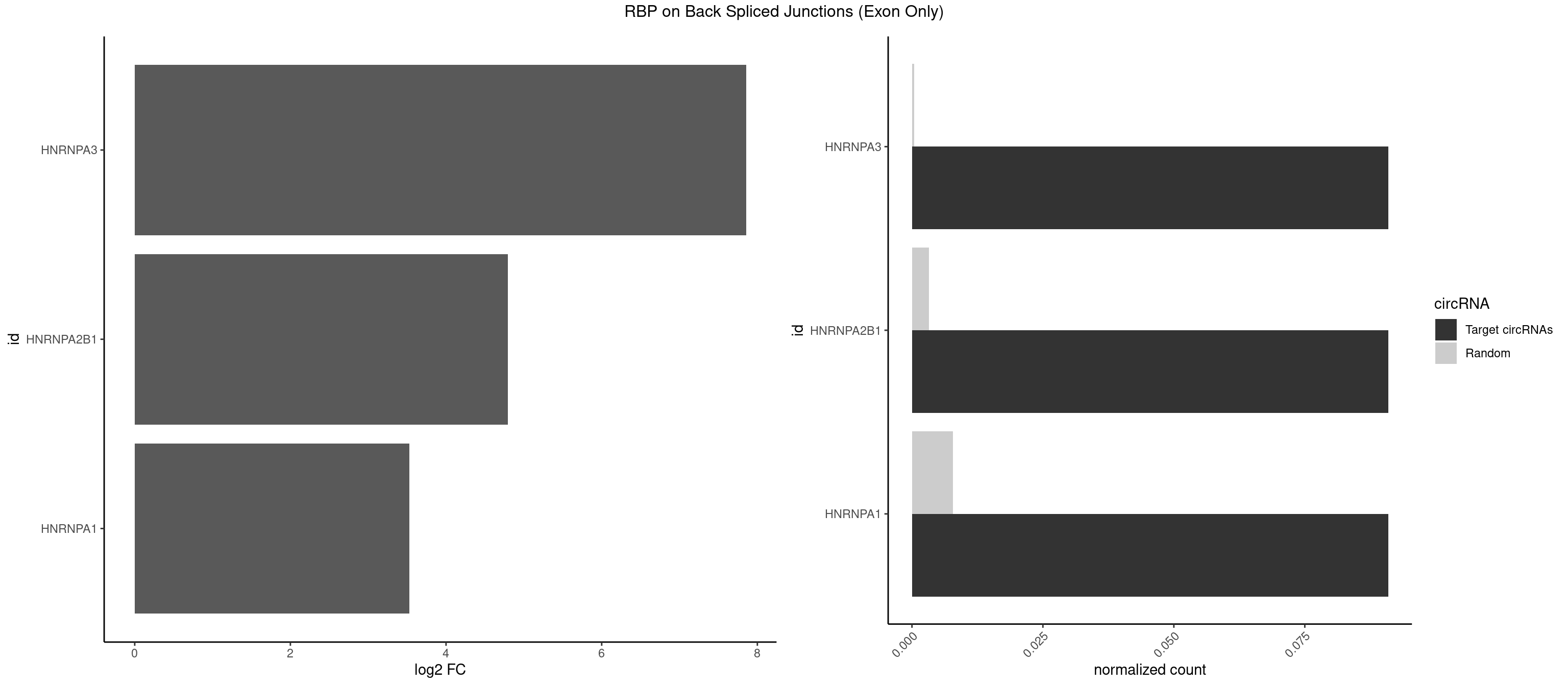
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA3 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GCCAAG | AGGAGC,CAAGGA,CCAAGG,GCCAAG |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | GCCAAG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.0078595756 | 3.531901 | GCCAAG | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
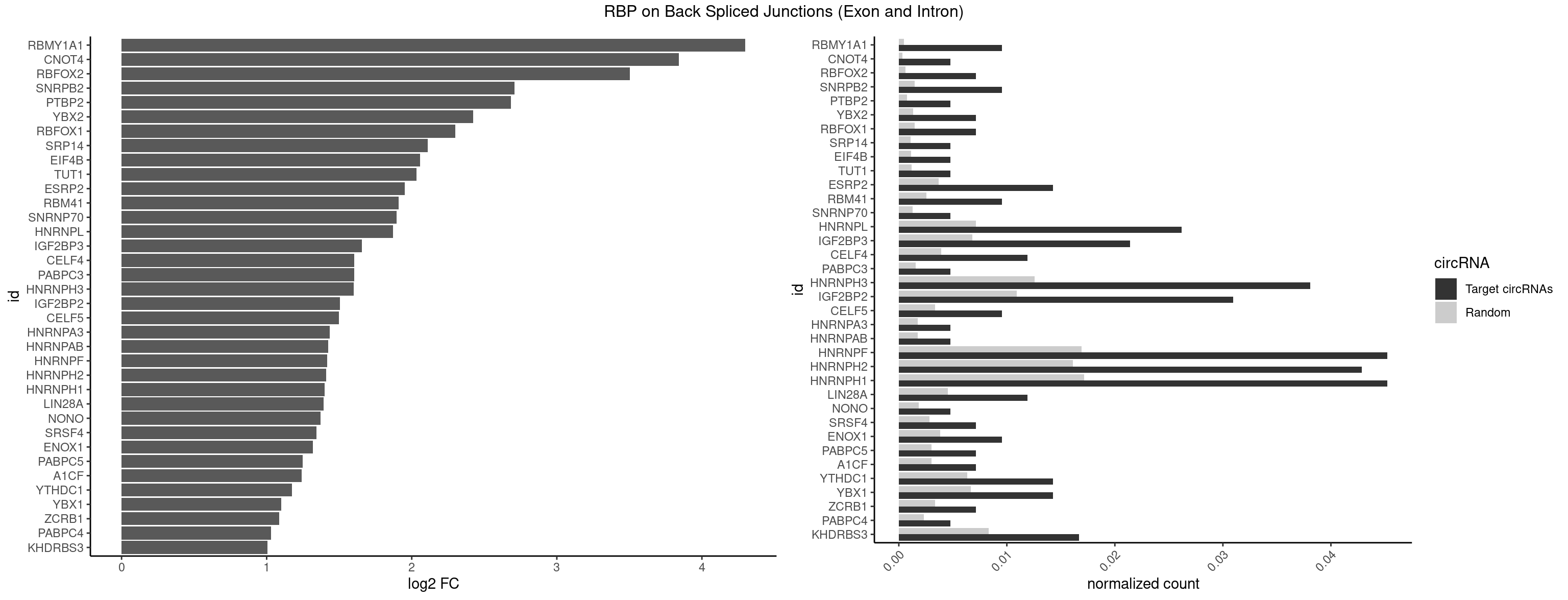
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 3 | 95 | 0.009523810 | 0.0004836759 | 4.299426 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGACUG | UGACUG,UGCAUG |
| SNRPB2 | 3 | 288 | 0.009523810 | 0.0014560661 | 2.709463 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAUC,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ESRP2 | 5 | 731 | 0.014285714 | 0.0036880290 | 1.953651 | GGGAAA,GGGAAG,GGGGAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | AUACAU,UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| HNRNPL | 10 | 1419 | 0.026190476 | 0.0071543732 | 1.872145 | AAACAA,AAACAC,AAAUAA,AAAUAC,AACACA,AAUAAA,AAUACA,ACAUAA,CACAAA,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| IGF2BP3 | 8 | 1348 | 0.021428571 | 0.0067966546 | 1.656639 | AAAACA,AAACAC,AAAUAC,AACACA,AAUACA,ACAAAC,CAAACA,CACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GGUGUG,GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPH3 | 15 | 2496 | 0.038095238 | 0.0125806127 | 1.598408 | AAGGGA,AGGGAA,AUGUGG,GAAGGG,GAGGGG,GGAAGG,GGAGGG,GGGAAG,GGGAGG,GGGCGU,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| IGF2BP2 | 12 | 2163 | 0.030952381 | 0.0109028617 | 1.505344 | AAAACA,AAACAC,AAAUAC,AACACA,AAUACA,ACAAAC,CAAAAC,CAAACA,CAACAC,CACUCA,CCACUC,GCAUUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPF | 18 | 3360 | 0.045238095 | 0.0169336961 | 1.417641 | AAGGGA,AGGAAG,AGGGAA,AUGUGG,GAAGGG,GAGGAA,GAGGGG,GGAAGG,GGAGGG,GGGAAG,GGGAGG,GGGGAA,UGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH2 | 17 | 3198 | 0.042857143 | 0.0161174929 | 1.410908 | AAGGGA,AGGGAA,AUGUGG,GAAGGG,GAGGGG,GGAAGG,GGAGGG,GGGAAG,GGGAGG,GGGCGU,GGGGAA,GGGGGA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 18 | 3406 | 0.045238095 | 0.0171654575 | 1.398030 | AAGGGA,AGGAAG,AGGGAA,AUGUGG,GAAGGG,GAGGAA,GAGGGG,GGAAGG,GGAGGG,GGGAAG,GGGAGG,GGGCGU,GGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGGG,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAGACA,AAUACA,AGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GACUGC,UCCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AACAUC,ACAUCU,CCACCA,CCCUGC,GAUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAAUAA,AAUAAA,AUAAAG,AUUAAA,UAAAUA,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.