circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000144040:-:2:73020238:73040931
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000144040:-:2:73020238:73040931 | ENSG00000144040 | ENST00000474528 | - | 2 | 73020239 | 73040931 | 186 | AGACGUCUCAGAGAGGCUGUGCAGCUGCUGGAGGACUAUAAGCAUGGGACCCUGCGCCCGGGGGUCACCAAUGAACAGCUCUGGAGUGCACAGAAAAUCAAGCAGGCUAUUCUACAUCCGGACACCAAUGAGAAGAUCUUCAUGCCAUUUAGAAUGUCAGGUUAUAUUCCUUUUGGGACGCCAAUUAGACGUCUCAGAGAGGCUGUGCAGCUGCUGGAGGACUAUAAGCAUGGGAC | circ |
| ENSG00000144040:-:2:73020238:73040931 | ENSG00000144040 | ENST00000474528 | - | 2 | 73020239 | 73040931 | 22 | GGACGCCAAUUAGACGUCUCAG | bsj |
| ENSG00000144040:-:2:73020238:73040931 | ENSG00000144040 | ENST00000474528 | - | 2 | 73040922 | 73041131 | 210 | UAGCUCUACUUCAUUUUUUAAACUGCCGAGUAAGAACAUUUAGGUGGUAUUUCUAUAUGAUUUUGUGUGUGUCCCAUGAACUGAGGGCAGGCCCAAAGCCUUGCACUGGCAGUAUUUGAUACUGAUGUAAUCCCACAAAAGAAAAAUUUGCGGGAGCCCUGGAUCUGUGCCCCUCAAUGUCUCUUUUCUUUCUUCUGCAGAGACGUCUCA | ie_up |
| ENSG00000144040:-:2:73020238:73040931 | ENSG00000144040 | ENST00000474528 | - | 2 | 73020039 | 73020248 | 210 | GACGCCAAUUGUAAGUGUUACCUUCAAAGGAUUUCCUUUUCUAAAAAAUUAUUUUAAAUGUCUAACUUUAUGUUAUUGCUCACGGGUAUUUGACUGAAUUGUUGAUUUAGGAUAAGUCAAUUCCUGGAGGGAAAUUACCAAAUAAAAUGAUAUGUAUUUCUUACCACAUCUCUUGAUGCUGAUUGGCAAACCUCUUUUCAUAAAAAGCCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
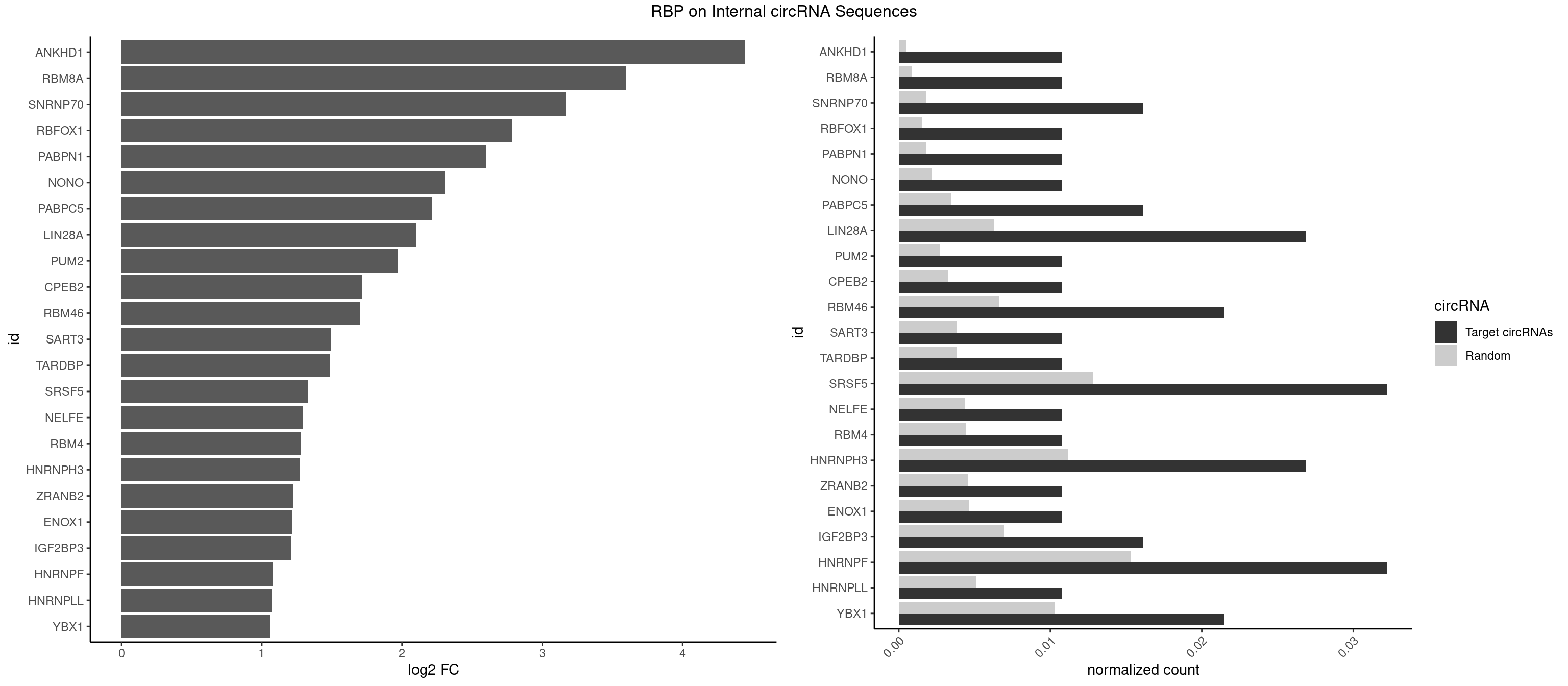
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 339 | 0.01075269 | 0.0004927293 | 4.447758 | AGACGU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM8A | 1 | 611 | 0.01075269 | 0.0008869128 | 3.599761 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| SNRNP70 | 2 | 1237 | 0.01612903 | 0.0017941145 | 3.168316 | AAUCAA,AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 1 | 1077 | 0.01075269 | 0.0015622419 | 2.783008 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPN1 | 1 | 1222 | 0.01075269 | 0.0017723764 | 2.600940 | AGAAGA | AAAAGA,AGAAGA |
| NONO | 1 | 1498 | 0.01075269 | 0.0021723567 | 2.307364 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 | 2 | 2400 | 0.01612903 | 0.0034795387 | 2.212692 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| LIN28A | 4 | 4315 | 0.02688172 | 0.0062547643 | 2.103598 | GGAGGA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PUM2 | 1 | 1890 | 0.01075269 | 0.0027404447 | 1.972215 | UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CPEB2 | 1 | 2261 | 0.01075269 | 0.0032780993 | 1.713766 | CCUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM46 | 3 | 4554 | 0.02150538 | 0.0066011240 | 1.703914 | AAUCAA,AAUGAA,AUCAAG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SART3 | 1 | 2634 | 0.01075269 | 0.0038186524 | 1.493562 | AGAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TARDBP | 1 | 2654 | 0.01075269 | 0.0038476365 | 1.482653 | GAAUGU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF5 | 5 | 8869 | 0.03225806 | 0.0128544391 | 1.327393 | AACAGC,AGAAGA,CACAGA,UACAUC,UGCAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| NELFE | 1 | 3028 | 0.01075269 | 0.0043896388 | 1.292523 | CUCUGG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 1 | 3065 | 0.01075269 | 0.0044432593 | 1.275007 | UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPH3 | 4 | 7688 | 0.02688172 | 0.0111429292 | 1.270497 | CGGGGG,GAAUGU,GGAGGA | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| ZRANB2 | 1 | 3173 | 0.01075269 | 0.0045997733 | 1.225063 | AGGUUA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ENOX1 | 1 | 3195 | 0.01075269 | 0.0046316558 | 1.215097 | CGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| IGF2BP3 | 2 | 4815 | 0.01612903 | 0.0069793662 | 1.208492 | AAAAUC,AAAUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPF | 5 | 10561 | 0.03225806 | 0.0153064921 | 1.075516 | AUGGGA,CGGGGG,GAAUGU,GGAGGA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPLL | 1 | 3534 | 0.01075269 | 0.0051229360 | 1.069655 | ACACCA | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| YBX1 | 3 | 7119 | 0.02150538 | 0.0103183321 | 1.059488 | ACACCA,CCCUGC,CCUGCG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
RBP on BSJ (Exon Only)
Plot
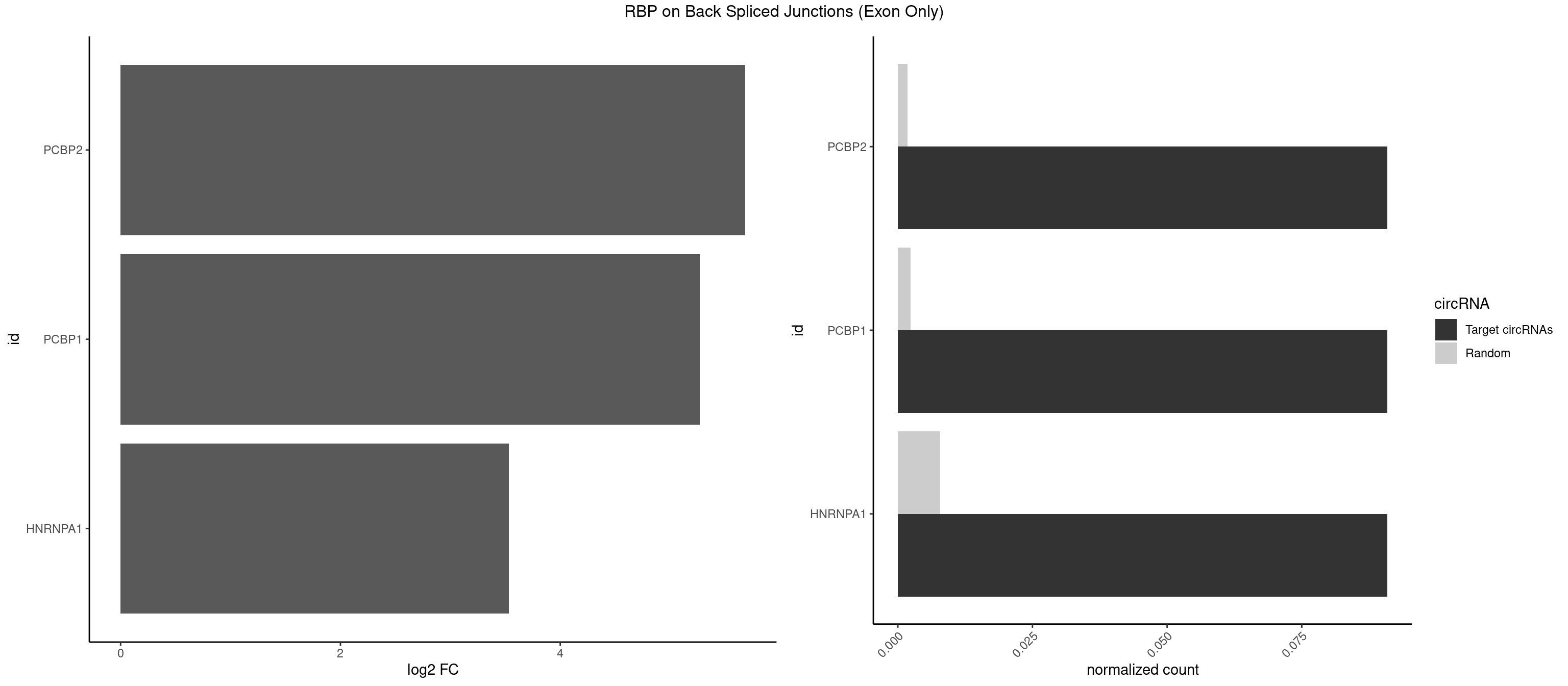
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PCBP2 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | CAAUUA | AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,AUUAAA,CCAUUC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CUCCCU,CUUCCA,UAACCC,UUAGAG,UUAGGA,UUAGGG |
| PCBP1 | 1 | 35 | 0.09090909 | 0.002357873 | 5.268867 | CAAUUA | AAAAAA,AAACCA,AAAUUA,AAAUUC,AAUUAA,ACCAAA,ACCCUC,AUUAAA,CAAACC,CAUACC,CCAACC,CCACCC,CCAUAC,CCAUUC,CCCACC,CCCCAC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,UUAGAG,UUAGGA,UUAGGG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | AUUAGA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
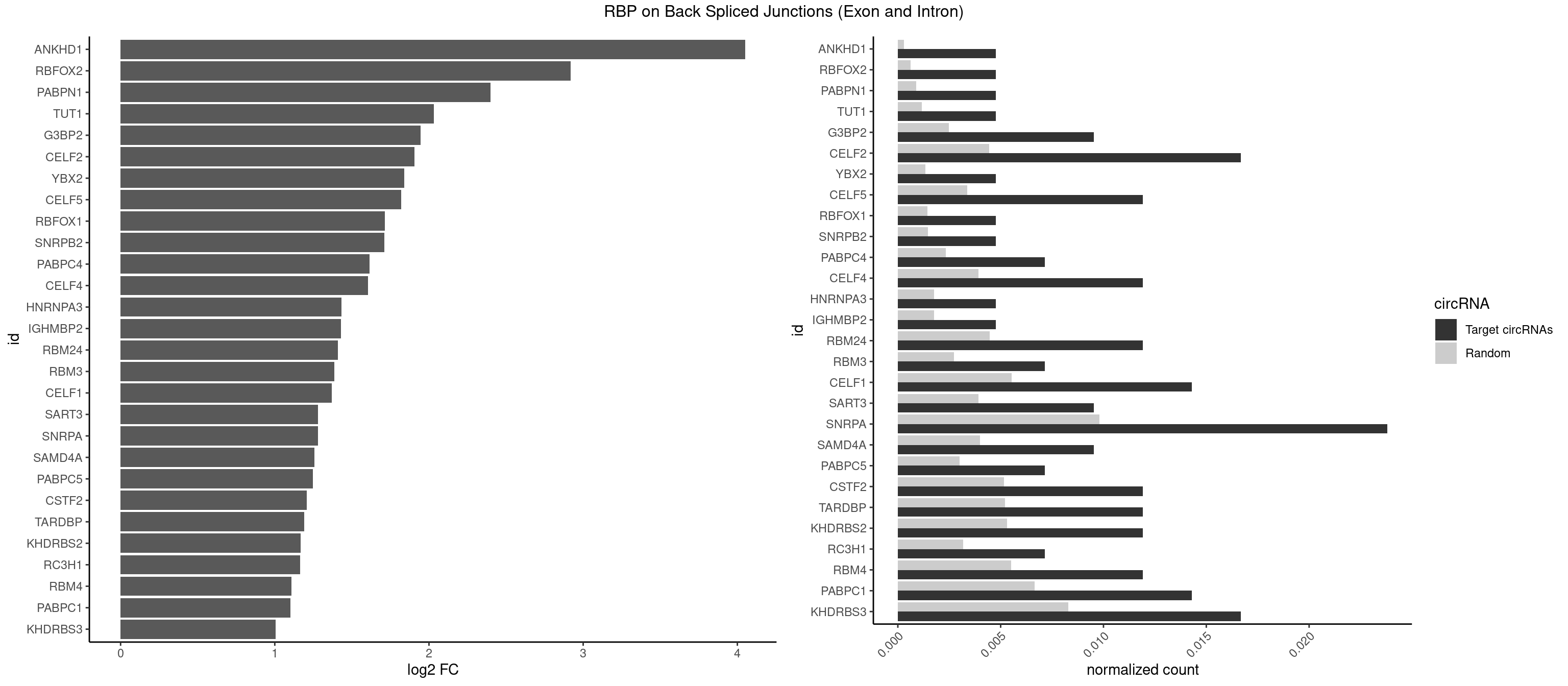
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | AGACGU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUA,AGGAUU,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CELF2 | 6 | 881 | 0.016666667 | 0.0044437727 | 1.907109 | GUGUGU,UAUGUU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | GAGACG,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | GUGUGU,UGUGUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SNRPA | 9 | 1947 | 0.023809524 | 0.0098145909 | 1.278539 | AUGCUG,AUUCCU,GCAGUA,GGGUAU,GUUACC,UUACCU,UUCCUG,UUGCAC,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CGGGUA,CUGGCA,GCGGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUGUGU,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CUCUUU,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA,AGAAAA,CAAACC,CAAAUA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAAUAA,AAUAAA,AUAAAA,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.