circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000064419:-:7:129014978:129018157
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000064419:-:7:129014978:129018157 | ENSG00000064419 | ENST00000627585 | - | 7 | 129014979 | 129018157 | 432 | GUUCAUGCAUGGGAGAUCUCAGACCAGUUGUUACAGAUCCGGCAGGAUGUGGAGUCAUGCUAUUUUGCUGCACAGACCAUGAAAAUGAAGAUUCAGACCUCAUUUUAUGAGCUCCCCACAGACUCUCAUGCCUCUUUACGGGACUCAUUGCUAACCCAUAUCCAGAACUUGAAAGACUUGUCACCUGUUAUUGUAACGCAGCUGGCUUUAGCAAUAGCAGAUCUUGCCCUACAGAUGCCUUCCUGGAAGGGAUGUGUGCAAACACUGGUGGAAAAAUACAGCAAUGAUGUGACUUCUUUGCCUUUUUUGCUGGAGAUCCUUACAGUGUUACCUGAAGAAGUACAUAGUCGUUCCUUACGAAUUGGAGCUAAUCGGCGCACAGAAAUUAUAGAAGAUUUGGCCUUCUACUCUAGUACAGUAGUAUCUCUAUUGGUUCAUGCAUGGGAGAUCUCAGACCAGUUGUUACAGAUCCGGCAGGAUGU | circ |
| ENSG00000064419:-:7:129014978:129018157 | ENSG00000064419 | ENST00000627585 | - | 7 | 129014979 | 129018157 | 22 | UAUCUCUAUUGGUUCAUGCAUG | bsj |
| ENSG00000064419:-:7:129014978:129018157 | ENSG00000064419 | ENST00000627585 | - | 7 | 129018148 | 129018357 | 210 | GAUUUUAGGUAAUUAUUUUGUUGAACAGGGCCUAUAACAAUGUAUAUUGAACUGUUGGCUGAAUUACUGGGUAAUUUAGCUUGUCAGAUUUUUCUUUCUUUAUACUUUGGGCUAAAUAUGUUUAUAUAAGUAUUGAGGUUAUGUCAUUAAAAUUAGAAAAGUAGUCUUCUUUAAGACAUCUUUGUCUAAUACGCUUUCAGGUUCAUGCAU | ie_up |
| ENSG00000064419:-:7:129014978:129018157 | ENSG00000064419 | ENST00000627585 | - | 7 | 129014779 | 129014988 | 210 | AUCUCUAUUGGUGAGUAAGUUUGAAAUACUAAGUUGCUGCAUAAAGGCAGAAAGCCAUGGCGGUUAUUUUGUAAUCAAUUGCAUUAUUGUGAAAUAGCUUUCUUUGCAAAAUUUAAUGAUGAUGCUUUGGUAUUUAUGUAACACCUUACCAUGACAACAUACUUUCAGAAACUAUGCUUUUUAAGUUUCUGUAUAAUCCUCUAAAGGUGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
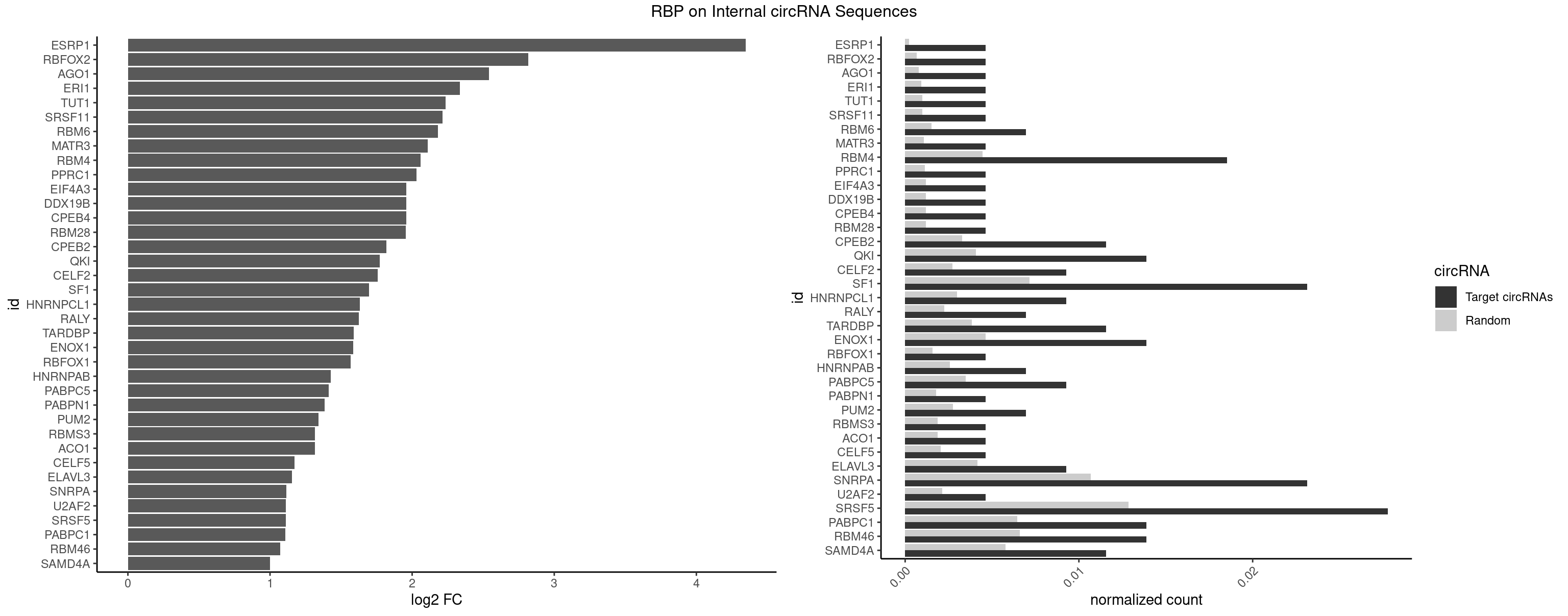
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.004629630 | 0.0002275250 | 4.346800 | AGGGAU | AGGGAU |
| RBFOX2 | 1 | 452 | 0.004629630 | 0.0006564894 | 2.818053 | UGCAUG | UGACUG,UGCAUG |
| AGO1 | 1 | 548 | 0.004629630 | 0.0007956130 | 2.540758 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ERI1 | 1 | 632 | 0.004629630 | 0.0009173461 | 2.335359 | UUCAGA | UUCAGA,UUUCAG |
| TUT1 | 1 | 678 | 0.004629630 | 0.0009840095 | 2.234153 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF11 | 1 | 688 | 0.004629630 | 0.0009985015 | 2.213060 | AAGAAG | AAGAAG |
| RBM6 | 2 | 1054 | 0.006944444 | 0.0015289102 | 2.183356 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 1 | 739 | 0.004629630 | 0.0010724109 | 2.110039 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM4 | 7 | 3065 | 0.018518519 | 0.0044432593 | 2.059278 | CCUCUU,CCUUCC,CCUUCU,CUCUUU,CUUCCU,CUUCUU,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PPRC1 | 1 | 780 | 0.004629630 | 0.0011318283 | 2.032242 | CGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| CPEB4 | 1 | 821 | 0.004629630 | 0.0011912456 | 1.958426 | UUUUUU | UUUUUU |
| DDX19B | 1 | 821 | 0.004629630 | 0.0011912456 | 1.958426 | UUUUUU | UUUUUU |
| EIF4A3 | 1 | 821 | 0.004629630 | 0.0011912456 | 1.958426 | UUUUUU | UUUUUU |
| RBM28 | 1 | 822 | 0.004629630 | 0.0011926949 | 1.956672 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CPEB2 | 4 | 2261 | 0.011574074 | 0.0032780993 | 1.819965 | CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| QKI | 5 | 2805 | 0.013888889 | 0.0040664663 | 1.772084 | ACUCAU,CUAACC,CUAAUC,CUACUC,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CELF2 | 3 | 1886 | 0.009259259 | 0.0027346479 | 1.759542 | AUGUGU,GUUGUU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SF1 | 9 | 4931 | 0.023148148 | 0.0071474739 | 1.695391 | ACAGAC,CACAGA,GCUAAC,UACGAA,UGCUAA,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| HNRNPCL1 | 3 | 2062 | 0.009259259 | 0.0029897078 | 1.630892 | CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RALY | 2 | 1553 | 0.006944444 | 0.0022520629 | 1.624612 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TARDBP | 4 | 2654 | 0.011574074 | 0.0038476365 | 1.588852 | GUUGUU,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ENOX1 | 5 | 3195 | 0.013888889 | 0.0046316558 | 1.584331 | AAUACA,AGUACA,AUACAG,GUACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBFOX1 | 1 | 1077 | 0.004629630 | 0.0015622419 | 1.567279 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPAB | 2 | 1782 | 0.006944444 | 0.0025839306 | 1.426292 | AAAGAC,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 | 3 | 2400 | 0.009259259 | 0.0034795387 | 1.412001 | AGAAAU,GAAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPN1 | 1 | 1222 | 0.004629630 | 0.0017723764 | 1.385212 | AGAAGA | AAAAGA,AGAAGA |
| PUM2 | 2 | 1890 | 0.006944444 | 0.0027404447 | 1.341449 | GUACAU,UACAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ACO1 | 1 | 1283 | 0.004629630 | 0.0018607779 | 1.314991 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 1 | 1283 | 0.004629630 | 0.0018607779 | 1.314991 | UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CELF5 | 1 | 1415 | 0.004629630 | 0.0020520728 | 1.173815 | UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL3 | 3 | 2867 | 0.009259259 | 0.0041563169 | 1.155591 | AUUUUA,UAUUUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| SNRPA | 9 | 7380 | 0.023148148 | 0.0106965744 | 1.113748 | AGUAGU,CAGUAG,GGAGAU,GUUACC,UACCUG,UGCACA,UUACCU,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| U2AF2 | 1 | 1477 | 0.004629630 | 0.0021419234 | 1.111990 | UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| SRSF5 | 11 | 8869 | 0.027777778 | 0.0128544391 | 1.111665 | AAGAAG,AGAAGA,CACAGA,CGCAGC,GAAGAA,UACAGA,UACAGC,UACAUA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| PABPC1 | 5 | 4443 | 0.013888889 | 0.0064402624 | 1.108740 | ACGAAU,CAAACA,CUAACC,CUAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM46 | 5 | 4554 | 0.013888889 | 0.0066011240 | 1.073148 | AAUGAA,AAUGAU,AUGAAA,AUGAAG,AUGAUG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SAMD4A | 4 | 3992 | 0.011574074 | 0.0057866714 | 1.000091 | CGGGAC,CUGGAA,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
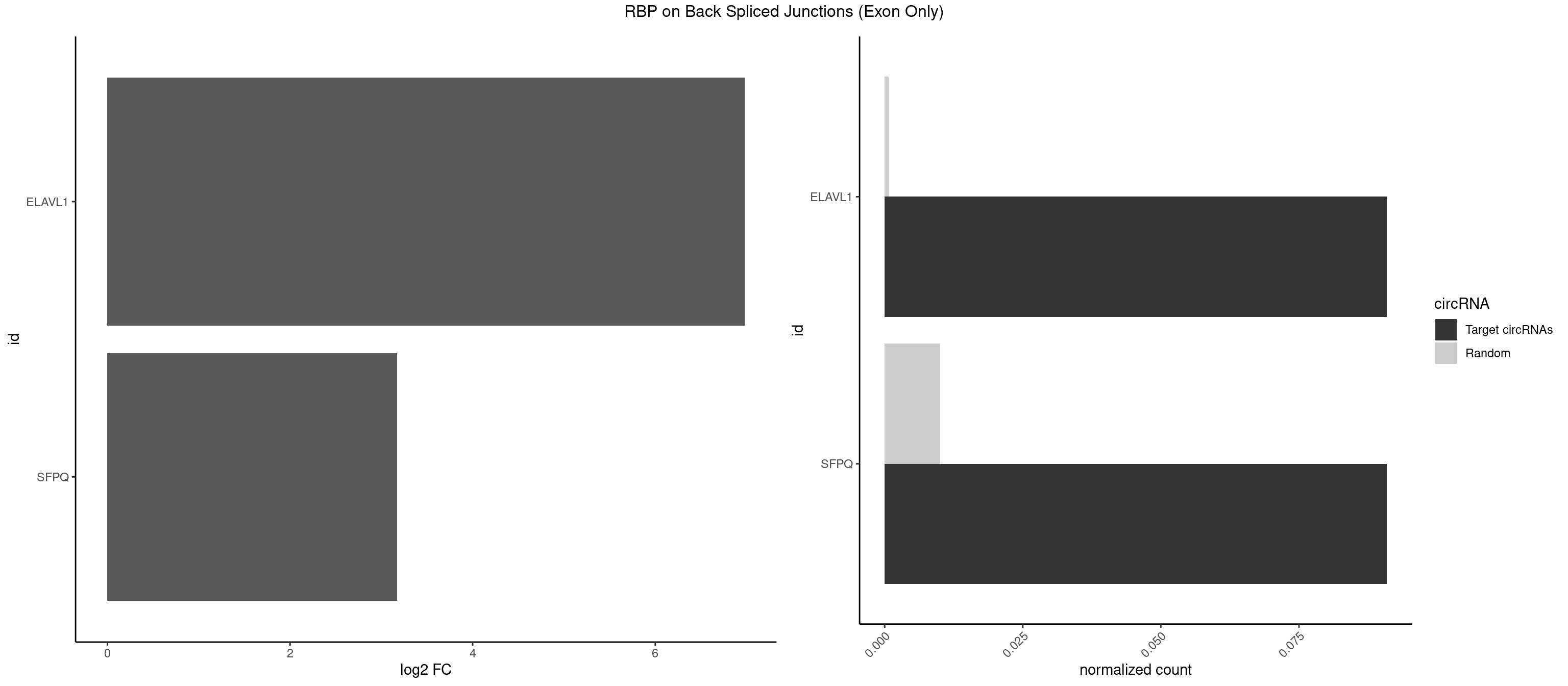
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ELAVL1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UUGGUU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | UUGGUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
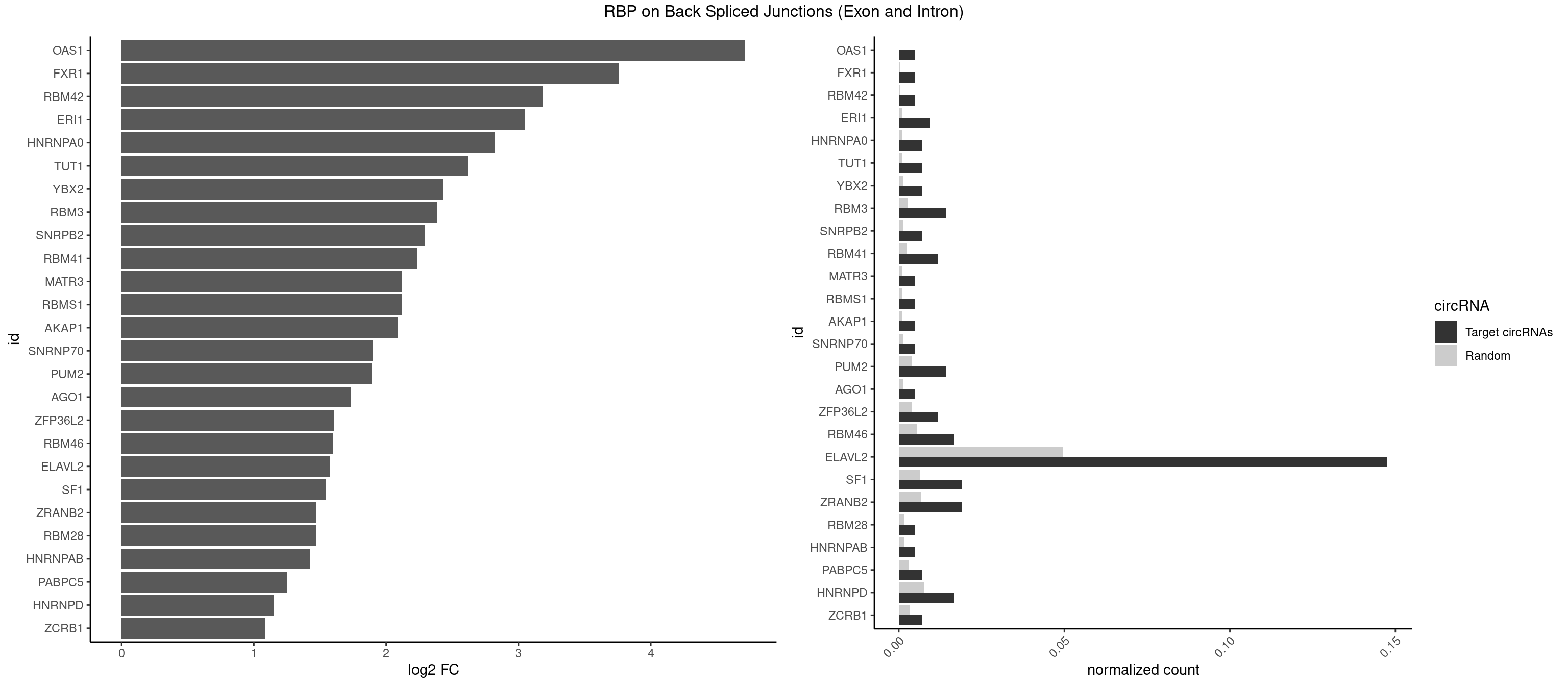
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| ERI1 | 3 | 228 | 0.009523810 | 0.0011537686 | 3.045185 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | ACAACA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM3 | 5 | 541 | 0.014285714 | 0.0027307537 | 2.387202 | AAACUA,AAUACG,AAUACU,AUACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | AUUGCA,GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | UAUAUA | AUAUAU,UAUAUA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | GUAUAU,UAAAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UAUUUA,UUAUUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUCAA,AAUGAU,AUCAAU,AUGAUG,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ELAVL2 | 61 | 9826 | 0.147619048 | 0.0495112858 | 1.576050 | AAUUUA,AUACUU,AUUAUU,AUUUAA,AUUUAG,AUUUAU,AUUUUA,AUUUUG,AUUUUU,CUUUAA,CUUUCU,CUUUUU,GAUUUU,GUAUUG,UAAGUU,UAAUUU,UACUUU,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUGA,UAUUGU,UAUUUA,UAUUUU,UUAAGU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCUUU,UUUAAG,UUUAAU,UUUAUA,UUUAUG,UUUCUU,UUUUAA,UUUUAG,UUUUUA,UUUUUC | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SF1 | 7 | 1294 | 0.019047619 | 0.0065245869 | 1.545652 | ACUAAG,AGUAAG,AUACUA,UAACAA,UACUAA,UAUACU,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ZRANB2 | 7 | 1360 | 0.019047619 | 0.0068571141 | 1.473937 | AAAGGU,AGGUAA,AGGUUA,GAGGUU,GGGUAA,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,AGAAAG | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AAUUUA,AUUUAU,UAUUUA,UUAUUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.