circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000147403:+:X:154399337:154399941
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000147403:+:X:154399337:154399941 | ENSG00000147403 | MSTRG.35107.3 | + | X | 154399338 | 154399941 | 306 | UUACCGGUAUUGUAAGAACAAGCCGUACCCAAAGUCUCGCUUCUGCCGAGGUGUCCCUGAUGCCAAGAUUCGCAUUUUUGACCUGGGGCGGAAAAAGGCAAAAGUGGAUGAGUUUCCGCUUUGUGGCCACAUGGUGUCAGAUGAAUAUGAGCAGCUGUCCUCUGAAGCCCUGGAGGCUGCCCGAAUUUGUGCCAAUAAGUACAUGGUAAAAAGUUGUGGCAAAGAUGGCUUCCAUAUCCGGGUGCGGCUCCACCCCUUCCACGUCAUCCGCAUCAACAAGAUGUUGUCCUGUGCUGGGGCUGACAGUUACCGGUAUUGUAAGAACAAGCCGUACCCAAAGUCUCGCUUCUGCCGAG | circ |
| ENSG00000147403:+:X:154399337:154399941 | ENSG00000147403 | MSTRG.35107.3 | + | X | 154399338 | 154399941 | 22 | GGGGCUGACAGUUACCGGUAUU | bsj |
| ENSG00000147403:+:X:154399337:154399941 | ENSG00000147403 | MSTRG.35107.3 | + | X | 154399138 | 154399347 | 210 | UUCUAGUCUUGGGUGUUGUCUGUGUUGCAGCAGCCAACUGUUGCUUUGUAGUUUAUUUCCCCGAAUGGAAACGGUUUAGAAGUGGACGUGCAUUCCCCACCCUUUUCCCGUCCCUCGUUUGGGUUGUUCCUUGAGGGGCAAAGUGCCUGUUGGGCUUUCUGUGAACCUCACCUAACCUGUGUUUUUUCACUCCCCUGCAGUUACCGGUAU | ie_up |
| ENSG00000147403:+:X:154399337:154399941 | ENSG00000147403 | MSTRG.35107.3 | + | X | 154399932 | 154400141 | 210 | GGGCUGACAGGUGAGCUUGGUCUGGGCCUUUUAAGGCAGUUGGAGUCUCUACAUUAUUAGGCUUGCAUUAUUUUAUCAGAGCAUAGAGGUGGCCCCAGUGACUCAGCCACUAUGGCUACUAGAAAAGCCAGGCUGGCAAGUGACUUUCAGUGGUCACUCAGGACCCCUUCCUGCAGAUACAAACAAAGCAUGAGUAAGUCUUAGCAAGCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
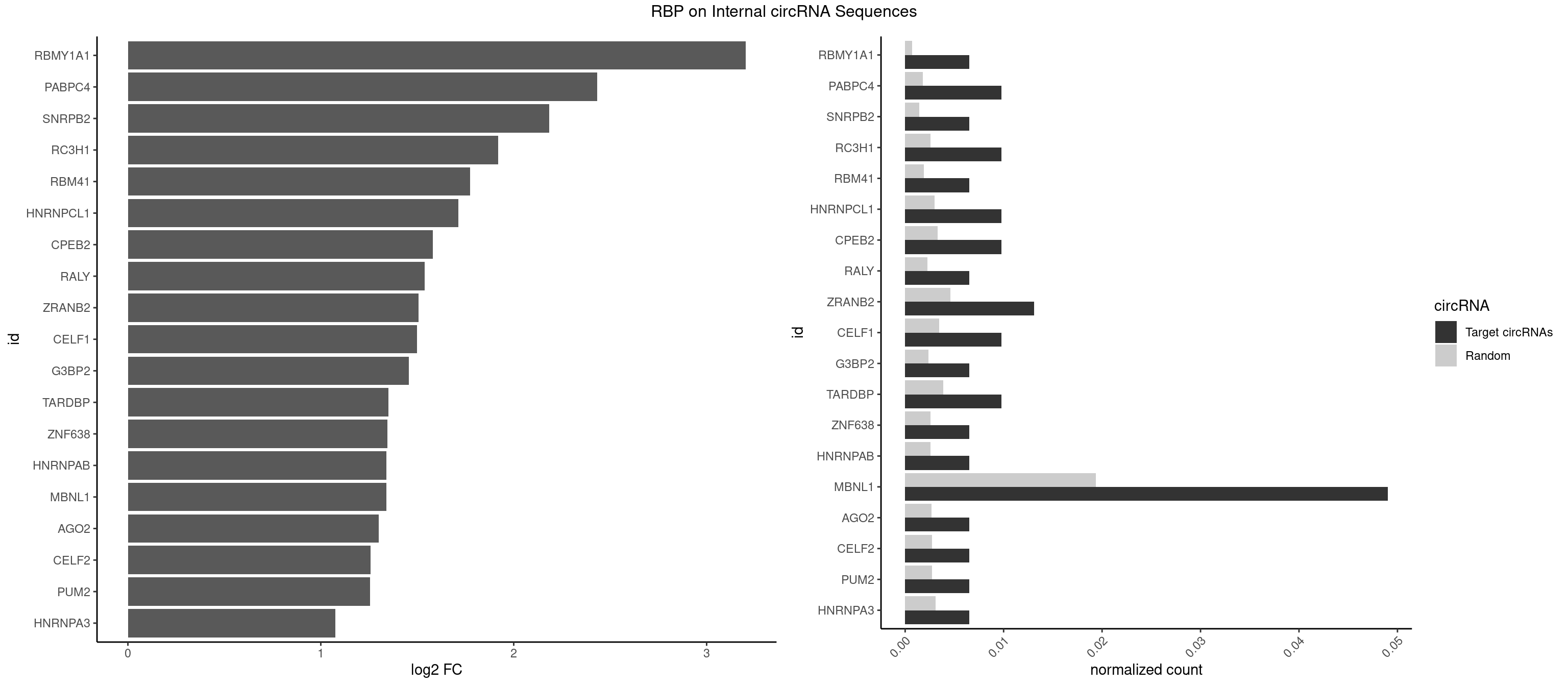
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 489 | 0.006535948 | 0.0007101099 | 3.202282 | ACAAGA | ACAAGA,CAAGAC |
| PABPC4 | 2 | 1251 | 0.009803922 | 0.0018144033 | 2.433864 | AAAAAG | AAAAAA,AAAAAG |
| SNRPB2 | 1 | 991 | 0.006535948 | 0.0014376103 | 2.184724 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RC3H1 | 2 | 1786 | 0.009803922 | 0.0025897275 | 1.920559 | CCCUUC,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM41 | 1 | 1318 | 0.006535948 | 0.0019115000 | 1.773691 | UACAUG | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPCL1 | 2 | 2062 | 0.009803922 | 0.0029897078 | 1.713354 | AUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 2 | 2261 | 0.009803922 | 0.0032780993 | 1.580499 | AUUUUU,CAUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RALY | 1 | 1553 | 0.006535948 | 0.0022520629 | 1.537149 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| ZRANB2 | 3 | 3173 | 0.013071895 | 0.0045997733 | 1.506834 | CGGUAU,GGUAAA,UGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| CELF1 | 2 | 2391 | 0.009803922 | 0.0034664959 | 1.499881 | GUUGUG,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| G3BP2 | 1 | 1644 | 0.006535948 | 0.0023839405 | 1.455048 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TARDBP | 2 | 2654 | 0.009803922 | 0.0038476365 | 1.349386 | GUUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 1 | 1773 | 0.006535948 | 0.0025708878 | 1.346130 | UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 1 | 1782 | 0.006535948 | 0.0025839306 | 1.338829 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MBNL1 | 14 | 13372 | 0.049019608 | 0.0193802045 | 1.338775 | AUGCCA,CCGCUU,CGCUUC,CGCUUU,CUCGCU,CUGCCC,CUGCCG,GUCUCG,GUGCUG,UCCGCU,UCGCUU,UCUCGC,UGCGGC,UUGUGC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| AGO2 | 1 | 1830 | 0.006535948 | 0.0026534924 | 1.300504 | AAAGUG | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CELF2 | 1 | 1886 | 0.006535948 | 0.0027346479 | 1.257041 | UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM2 | 1 | 1890 | 0.006535948 | 0.0027404447 | 1.253986 | GUACAU | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPA3 | 1 | 2140 | 0.006535948 | 0.0031027457 | 1.074851 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
RBP on BSJ (Exon Only)
Plot
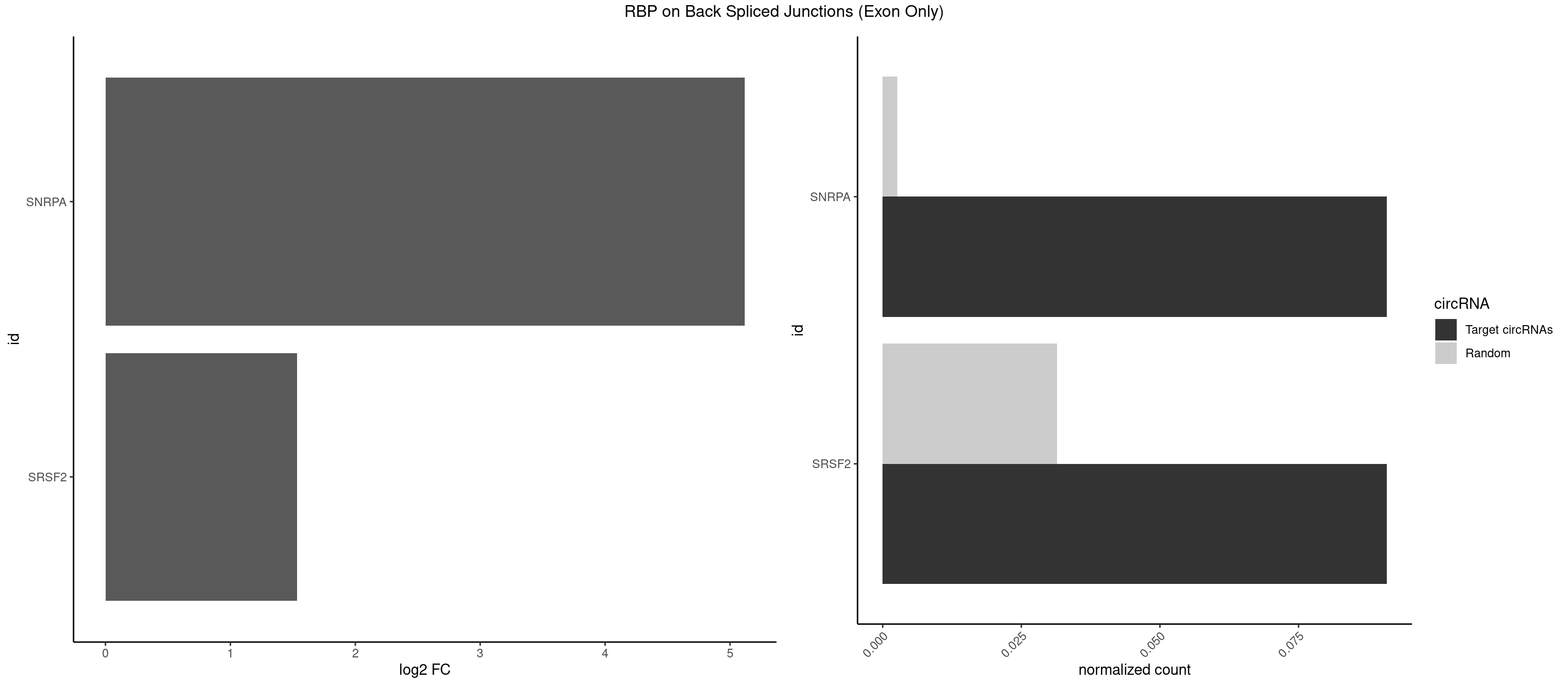
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPA | 1 | 39 | 0.09090909 | 0.002619859 | 5.116864 | GUUACC | AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,GAGCAG,GAUACC,GCAGUA,GGAGAU,GGGUAU,GUAGGC,GUAGUG,UAUGCU,UCCUGC,UGCACG,UUCCUG,UUGCAC |
| SRSF2 | 1 | 479 | 0.09090909 | 0.031438302 | 1.531901 | GUUACC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
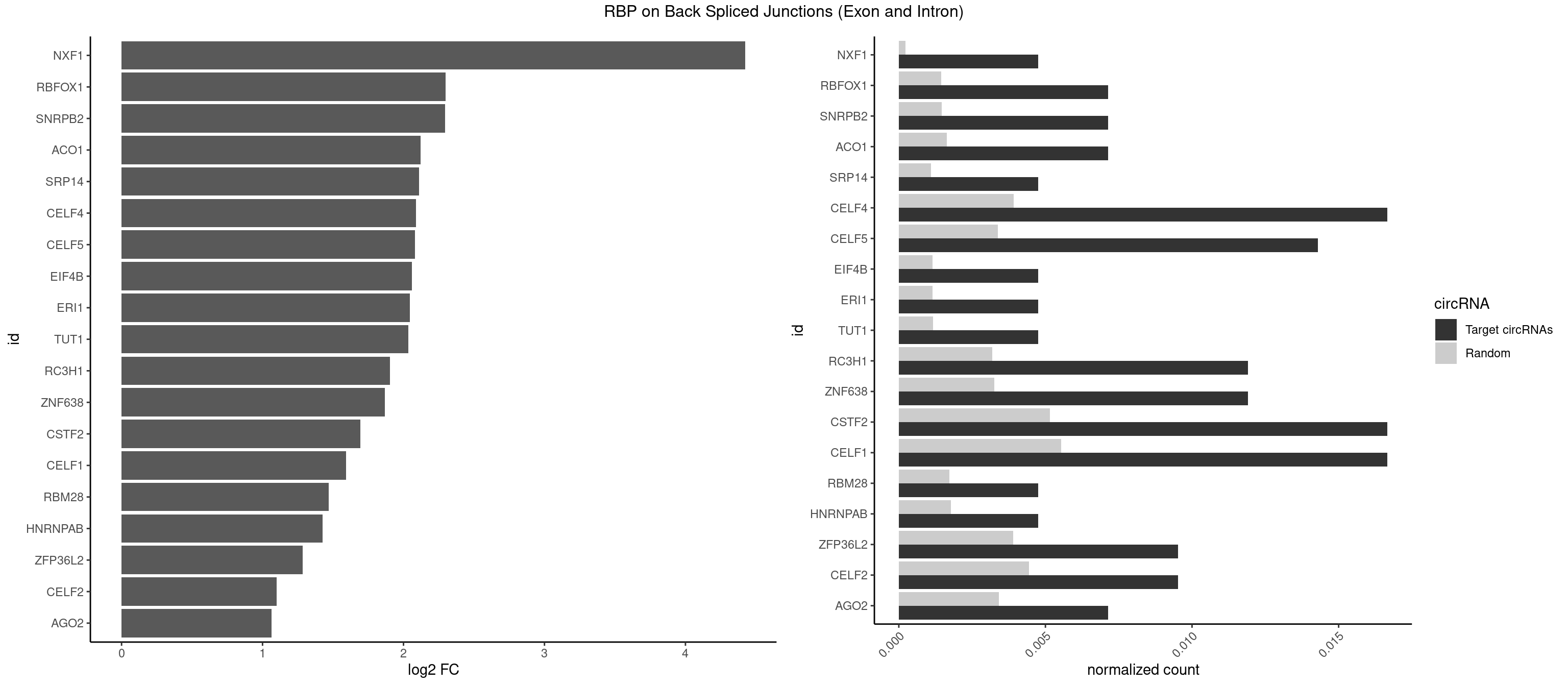
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF4 | 6 | 776 | 0.016666667 | 0.0039147521 | 2.089973 | GGUGUU,GUGUUG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUUG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CCCUUC,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | GGUUGU,GUUGUU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CSTF2 | 6 | 1022 | 0.016666667 | 0.0051541717 | 1.693153 | GUGUUG,GUGUUU,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF1 | 6 | 1097 | 0.016666667 | 0.0055320435 | 1.591081 | GUGUUG,UGUCUG,UGUGUU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUCUGU,GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.