circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000117262:+:1:145616233:145647267
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000117262:+:1:145616233:145647267 | ENSG00000117262 | MSTRG.2007.5 | + | 1 | 145616234 | 145647267 | 944 | AUACUAUUUUUUGGAUUUGGGUGGCUUUUCUUCAUGCGCCAAUUGUUUAAAGACUAUGAGAUACGUCAGUAUGUUGUACAGGUGAUCUUCUCCGUGACGUUUGCAUUUUCUUGCACCAUGUUUGAGCUCAUCAUCUUUGAAAUCUUAGGAGUAUUGAAUAGCAGCUCCCGUUAUUUUCACUGGAAAAUGAACCUGUGUGUAAUUCUGCUGAUCCUGGUUUUCAUGGUGCCUUUUUACAUUGGCUAUUUUAUUGUGAGCAAUAUCCGACUACUGCAUAAACAACGACUGCUUUUUUCCUGUCUCUUAUGGCUGACCUUUAUGUAUUUCUUCUGGAAACUAGGAGAUCCCUUUCCCAUUCUCAGCCCAAAACAUGGGAUCUUAUCCAUAGAACAGCUCAUCAGCCGGGUUGGUGUGAUUGGAGUGACUCUCAUGGCUCUUCUUUCUGGAUUUGGUGCUGUCAACUGCCCAUACACUUACAUGUCUUACUUCCUCAGGAAUGUGACUGACACGGAUAUUCUAGCCCUGGAACGGCGACUGCUGCAAACCAUGGAUAUGAUCAUAAGCAAAAAGAAAAGGAUGGCAAUGGCACGGAGAACAAUGUUCCAGAAGGGGGAAGUGCAUAACAAACCAUCAGGUUUCUGGGGAAUGAUAAAAAGUGUUACCACUUCAGCAUCAGGAAGUGAAACUUCACUUUCCUUAUGUGCAAAAUGGAUAACACCAUGGCAUAAGACAAUCCCAUGAAGAUAAGAUAUAAAAGAAGAUAAUGUUUGAAAAGAAAGAAGGAAGUGGAUGCUUUGGAAGAAUUAAGCAGGCAGCUUUUUCUGGAAACAGCUGAUCUAUAUGCUACCAAGGAGAGAAUAGAAUACUCCAAAACCUUCAAGGGGAAAUAUUUUAAUUUUCUUGGUUACUUUUUCUCUAUUUACUGUGUUUGGAAAAUUUUCAUGAUACUAUUUUUUGGAUUUGGGUGGCUUUUCUUCAUGCGCCAAUUGUUUAA | circ |
| ENSG00000117262:+:1:145616233:145647267 | ENSG00000117262 | MSTRG.2007.5 | + | 1 | 145616234 | 145647267 | 22 | AAAUUUUCAUGAUACUAUUUUU | bsj |
| ENSG00000117262:+:1:145616233:145647267 | ENSG00000117262 | MSTRG.2007.5 | + | 1 | 145616034 | 145616243 | 210 | CUUCAAAACUCCAAGGGCAUUUAAAAAUUUAUUUCCAAUGUCAUGAGCUUUAAUAAUAGGUCUUAUAAGAGUGGAUCUUAUCUUUAAUCCCUGAAAUAAGUUAACCGUCUAGUUAGCAGACCUUUUAUCAUAAAAGAGUUUCUAUAAAACAUUUCUCAAAAGAAAAUAUGUAUUGACAUUCUAUUUUCUUUCUCCUCCAGAUACUAUUUU | ie_up |
| ENSG00000117262:+:1:145616233:145647267 | ENSG00000117262 | MSTRG.2007.5 | + | 1 | 145647258 | 145647467 | 210 | AAUUUUCAUGGUAAGUAUGUUAUUUUUAAUUAUCAGAGUUUAGAUGUUUUUUCCUGGUAGUGGCAGGGAGAUAAGAGUGGAUUUGACUGAUAUAUUGAACUAAAUUCCAAAUGUGAUUGCAUCAACAGAGAGCAUGAUCUUCCACAUCUCUGGGCUUCAUUCGUUAUCCUCUCUCCUUUUUAGAUUACAUAGAUAAGAAAGGUUGCAUUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
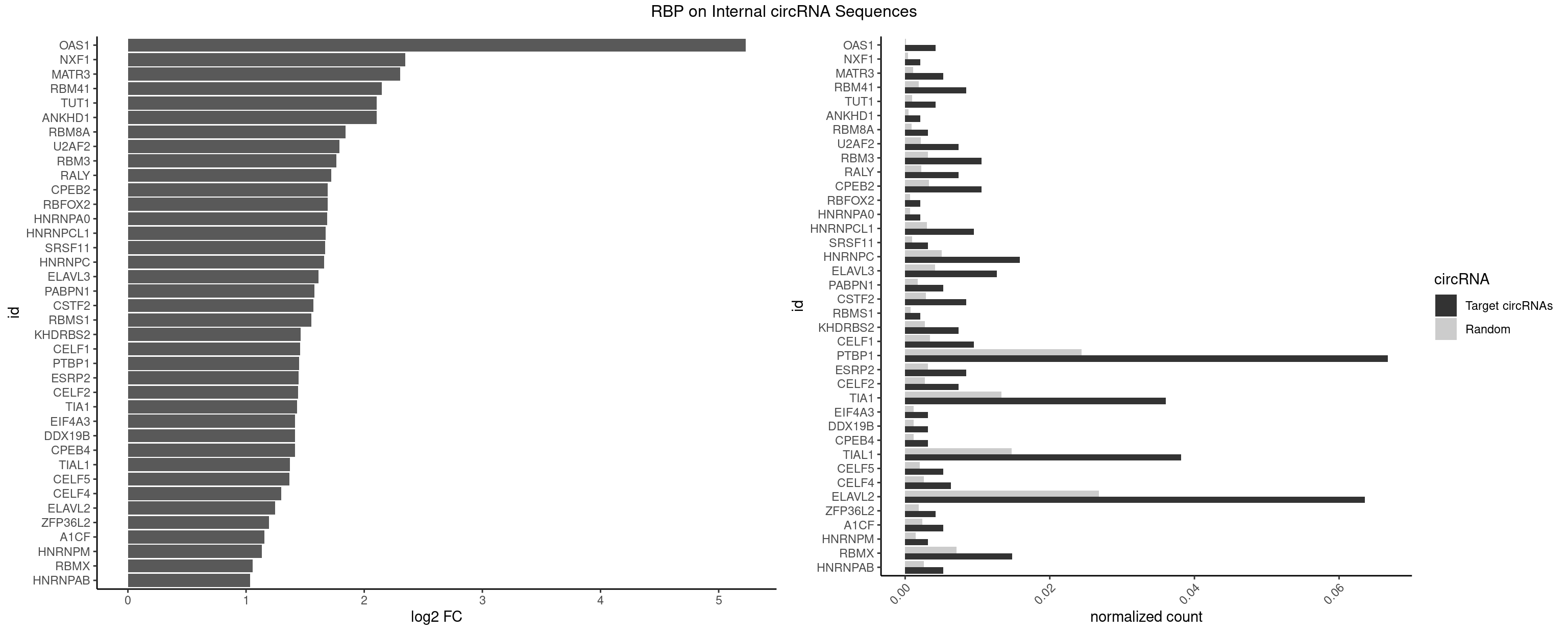
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 3 | 77 | 0.004237288 | 0.0001130379 | 5.228263 | GCAUAA | GCAUAA |
| NXF1 | 1 | 286 | 0.002118644 | 0.0004159215 | 2.348758 | AACCUG | AACCUG |
| MATR3 | 4 | 739 | 0.005296610 | 0.0010724109 | 2.304212 | AAUCUU,AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM41 | 7 | 1318 | 0.008474576 | 0.0019115000 | 2.148436 | UACAUG,UACAUU,UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| TUT1 | 3 | 678 | 0.004237288 | 0.0009840095 | 2.106397 | AAUACU,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ANKHD1 | 1 | 339 | 0.002118644 | 0.0004927293 | 2.104274 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM8A | 2 | 611 | 0.003177966 | 0.0008869128 | 1.841240 | AUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| U2AF2 | 6 | 1477 | 0.007415254 | 0.0021419234 | 1.791589 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM3 | 9 | 2152 | 0.010593220 | 0.0031201361 | 1.763460 | AAACUA,AAGACU,AAUACU,AGACUA,AUACUA,GAAACU,GAUACG,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| RALY | 6 | 1553 | 0.007415254 | 0.0022520629 | 1.719249 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| CPEB2 | 9 | 2261 | 0.010593220 | 0.0032780993 | 1.692210 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBFOX2 | 1 | 452 | 0.002118644 | 0.0006564894 | 1.690298 | UGACUG | UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.002118644 | 0.0006579386 | 1.687116 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| HNRNPCL1 | 8 | 2062 | 0.009533898 | 0.0029897078 | 1.673062 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SRSF11 | 2 | 688 | 0.003177966 | 0.0009985015 | 1.670267 | AAGAAG | AAGAAG |
| HNRNPC | 14 | 3471 | 0.015889831 | 0.0050316361 | 1.659004 | AUUUUU,CUUUUU,GGAUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 11 | 2867 | 0.012711864 | 0.0041563169 | 1.612798 | AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PABPN1 | 4 | 1222 | 0.005296610 | 0.0017723764 | 1.579384 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| CSTF2 | 7 | 1967 | 0.008474576 | 0.0028520334 | 1.571150 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBMS1 | 1 | 497 | 0.002118644 | 0.0007217036 | 1.553663 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| KHDRBS2 | 6 | 1858 | 0.007415254 | 0.0026940701 | 1.460709 | AUAAAA,AUAAAC,GAUAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CELF1 | 8 | 2391 | 0.009533898 | 0.0034664959 | 1.459588 | CUGUCU,GUGUGU,UGUGUG,UGUGUU,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PTBP1 | 62 | 16854 | 0.066737288 | 0.0244263326 | 1.450056 | ACUUUC,ACUUUU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCUUUC,CCUUUU,CUCUUA,CUCUUC,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GUCUUA,UACUUU,UAUUUU,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUAUUU,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| ESRP2 | 7 | 2150 | 0.008474576 | 0.0031172377 | 1.442873 | GGGAAA,GGGAAG,GGGAAU,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| CELF2 | 6 | 1886 | 0.007415254 | 0.0027346479 | 1.439141 | GUAUGU,GUGUGU,UAUGUG,UAUGUU,UGUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| TIA1 | 33 | 9206 | 0.036016949 | 0.0133428208 | 1.432612 | AUUUUC,AUUUUU,CCUUUC,CUUUUC,CUUUUU,GUUUUC,UAUUUU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUGG,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CPEB4 | 2 | 821 | 0.003177966 | 0.0011912456 | 1.415633 | UUUUUU | UUUUUU |
| DDX19B | 2 | 821 | 0.003177966 | 0.0011912456 | 1.415633 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 821 | 0.003177966 | 0.0011912456 | 1.415633 | UUUUUU | UUUUUU |
| TIAL1 | 35 | 10182 | 0.038135593 | 0.0147572438 | 1.369715 | AAAUUU,AAUUUU,AUUUUA,AUUUUC,AUUUUU,CUUUUC,CUUUUU,GUUUUC,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF5 | 4 | 1415 | 0.005296610 | 0.0020520728 | 1.367987 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 5 | 1782 | 0.006355932 | 0.0025839306 | 1.298536 | GGUGUG,GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL2 | 59 | 18468 | 0.063559322 | 0.0267653478 | 1.247737 | AAUUUU,AUAUUU,AUUUAC,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUUCU,CUUUUU,GUAUUG,GUUUUC,UAAUUU,UACUUU,UAUGUU,UAUUGA,UAUUGU,UAUUUA,UAUUUU,UCUUAU,UGUAUU,UUAAUU,UUACUU,UUAUGU,UUAUUG,UUAUUU,UUCUUU,UUUAAU,UUUACU,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZFP36L2 | 3 | 1277 | 0.004237288 | 0.0018520827 | 1.193993 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| A1CF | 4 | 1642 | 0.005296610 | 0.0023810421 | 1.153476 | CAGUAU,UCAGUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPM | 2 | 999 | 0.003177966 | 0.0014492040 | 1.132843 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBMX | 13 | 4925 | 0.014830508 | 0.0071387787 | 1.054819 | AAGAAG,AAGGAA,AAGUGU,AGAAGG,AGGAAG,AGUGUU,AUCCCA,GAAGGA,UAACAA,UAAGAC | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| HNRNPAB | 4 | 1782 | 0.005296610 | 0.0025839306 | 1.035502 | AAAGAC,AAGACA,AGACAA,AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
RBP on BSJ (Exon Only)
Plot
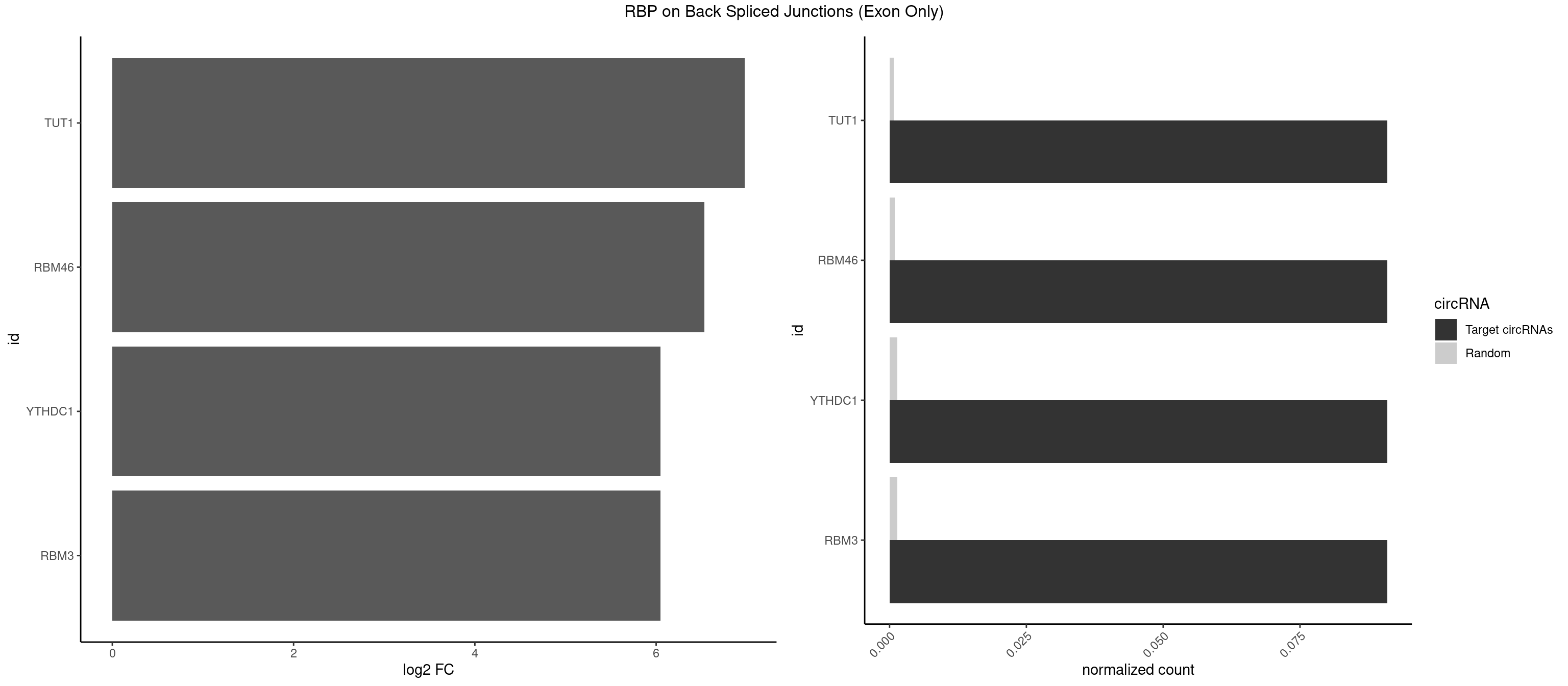
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TUT1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | AUGAUA | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
| RBM3 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | GAUACU | AAAACG,AAAACU,AAGACU,AAUACG,AAUACU,AUACUA,GAAACU,GAGACG,GAGACU,GAUACU |
| YTHDC1 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | UGAUAC | GAAUAC,GAGUAC,GCCUGC,GGCUGC,GGGUAC,UAAUAC,UAAUGC,UCAUAC,UCCUGC,UCGUGC,UGAUAC,UGCUGC,UGGUGC |
RBP on BSJ (Exon and Intron)
Plot
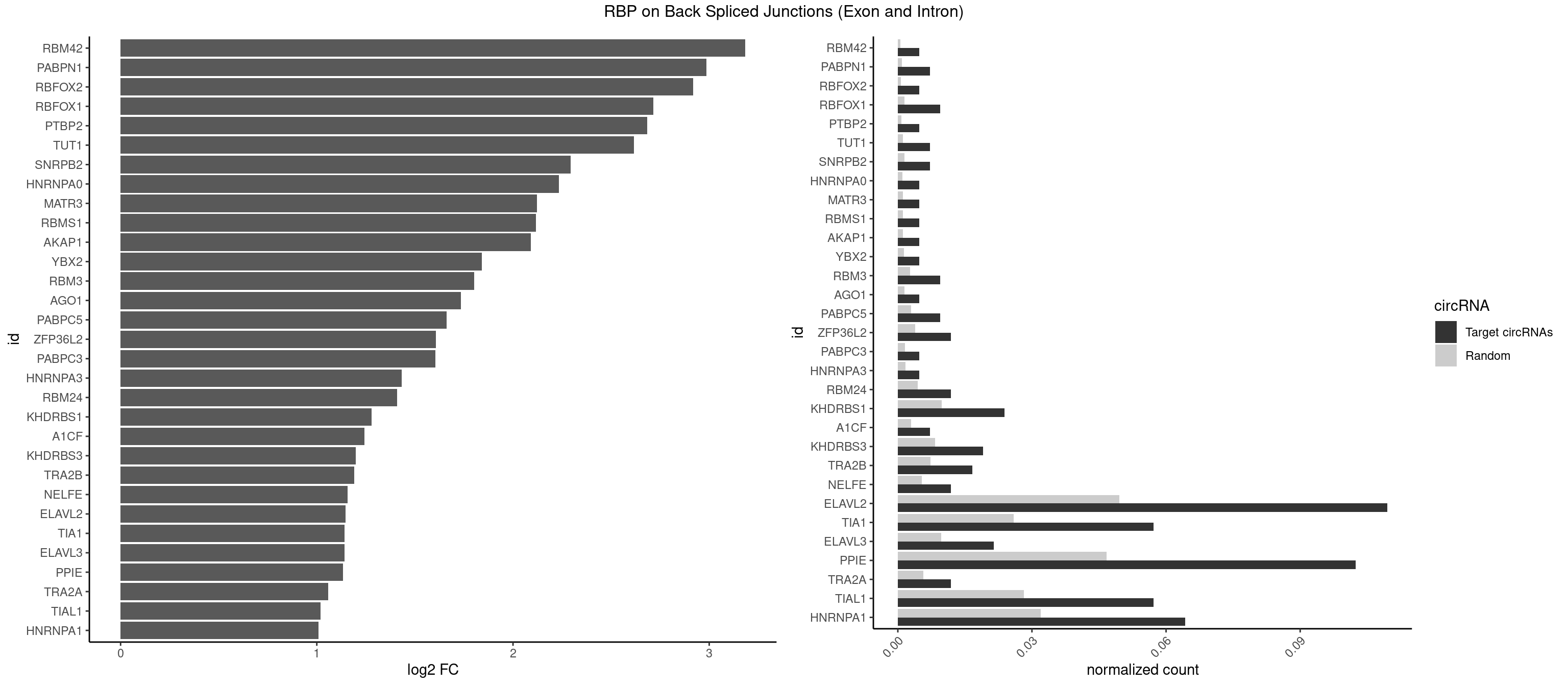
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGA,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | AUUGCA,GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM3 | 3 | 541 | 0.009523810 | 0.0027307537 | 1.802240 | AAAACU,AUACUA,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,AGAAAG,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | CCAAGG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,GAGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUAAAA,AUUUAA,CUAAAU,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | UAAUUA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AAAUAA,AGAUAA,AUAAAA,UAAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAUAAG,AUAAGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUCU,CUCUGG,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ELAVL2 | 45 | 9826 | 0.109523810 | 0.0495112858 | 1.145415 | AAUUUA,AAUUUU,AUUUAA,AUUUAU,AUUUUC,AUUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GUAUUG,GUUUUU,UAAGUU,UAGUUA,UAUAUU,UAUGUU,UAUUGA,UAUUUU,UCUUAU,UGUAUU,UUAAUU,UUAUUU,UUCAUU,UUCUUU,UUUAAU,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUC,AUUUUU,CUCCUU,CUUUUA,CUUUUU,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUAU,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PPIE | 42 | 9262 | 0.102380952 | 0.0466696896 | 1.133390 | AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUU,AAUAAU,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAUA,AUAUAU,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAAUU,UAAUAA,UAAUUA,UAUAAA,UAUAUU,UAUUUU,UUAAAA,UUAAUA,UUAAUU,UUAUAA,UUAUUU,UUUAAA,UUUAAU,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAAGAA,AAGAAA,AGAAAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TIAL1 | 23 | 5597 | 0.057142857 | 0.0282043531 | 1.018655 | AAAUUU,AAUUUU,AUUUUC,AUUUUU,CUUUUA,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1 | 26 | 6340 | 0.064285714 | 0.0319478033 | 1.008781 | AAAGAG,AAUUUA,AGAUUA,AGGGCA,AGUUAG,AUUUAA,AUUUAU,CAGGGA,CCAAGG,GAGUGG,GGCAGG,GUAAGU,GUUUAG,UAAGUA,UAGAUU,UAGGUC,UAGUUA,UUAGAU,UUAUUU,UUUAGA,UUUAUU,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.