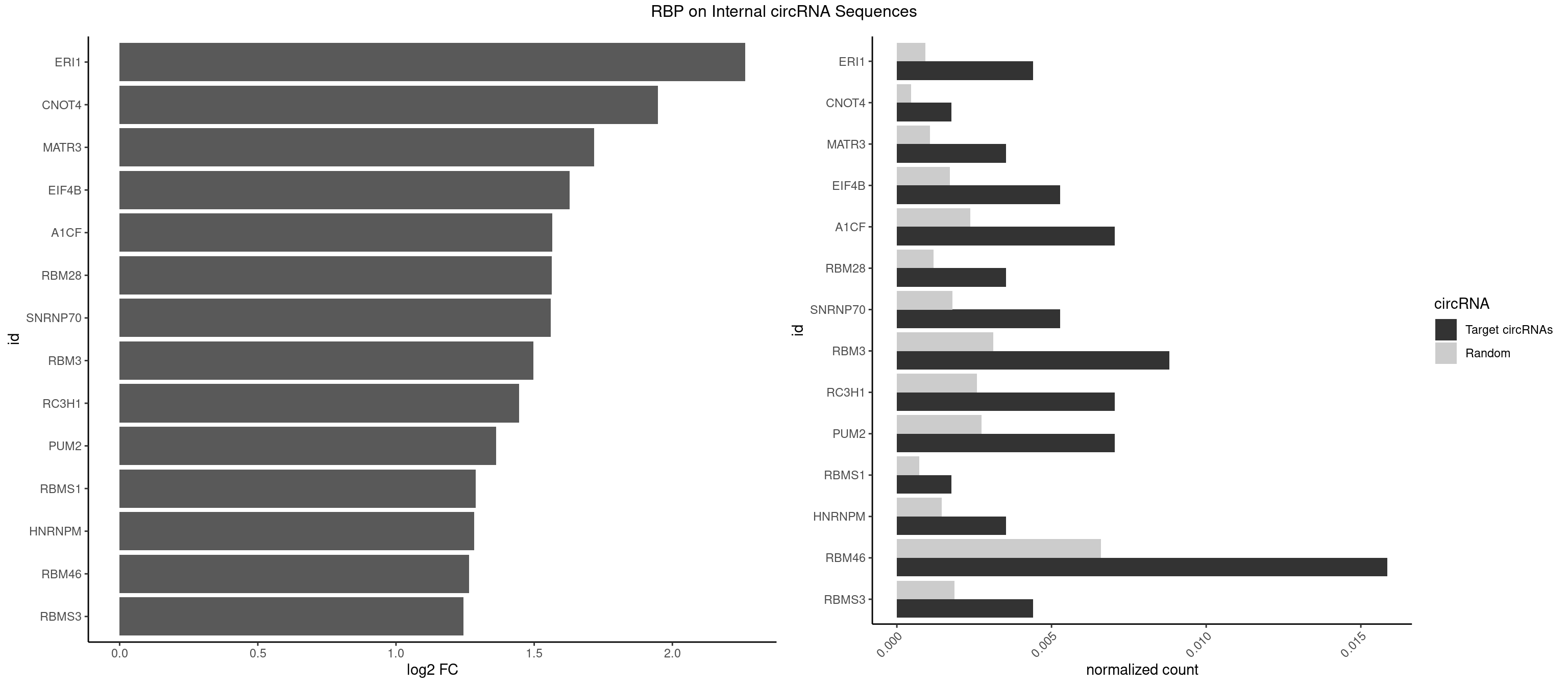
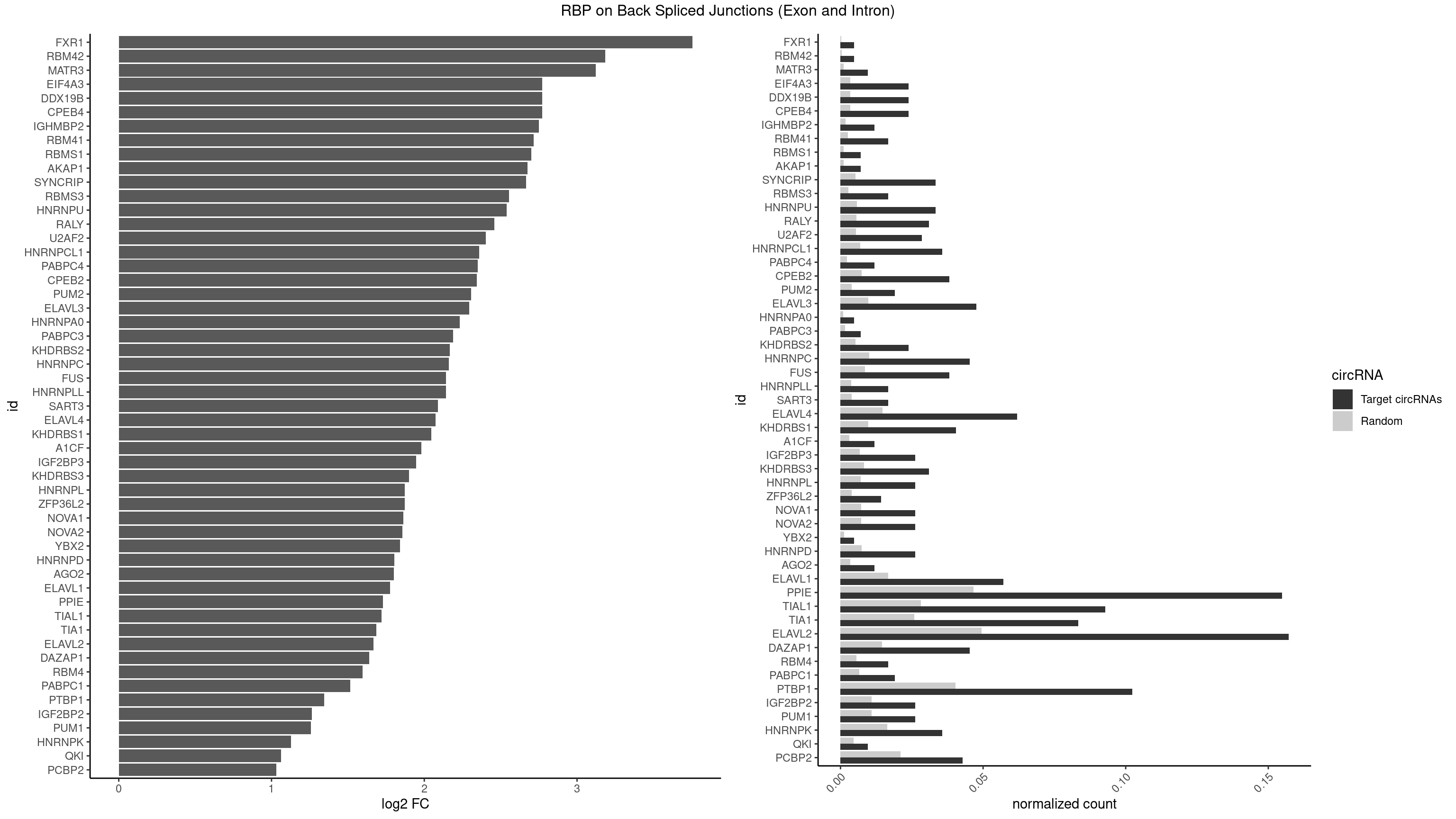
| FXR1 |
1 |
69 |
0.004761905 |
0.0003526804 |
3.755106 |
AUGACA |
ACGACA,ACGACG,AUGACA,AUGACG |
| RBM42 |
1 |
103 |
0.004761905 |
0.0005239823 |
3.183949 |
AACUAC |
AACUAA,AACUAC,ACUAAG,ACUACG |
| MATR3 |
3 |
216 |
0.009523810 |
0.0010933091 |
3.122837 |
AUCUUA,AUCUUG,CAUCUU |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CPEB4 |
9 |
691 |
0.023809524 |
0.0034864974 |
2.771688 |
UUUUUU |
UUUUUU |
| DDX19B |
9 |
691 |
0.023809524 |
0.0034864974 |
2.771688 |
UUUUUU |
UUUUUU |
| EIF4A3 |
9 |
691 |
0.023809524 |
0.0034864974 |
2.771688 |
UUUUUU |
UUUUUU |
| IGHMBP2 |
4 |
350 |
0.011904762 |
0.0017684401 |
2.750989 |
AAAAAA |
AAAAAA |
| RBM41 |
6 |
502 |
0.016666667 |
0.0025342604 |
2.717329 |
AUACAU,UACAUU,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBMS1 |
2 |
217 |
0.007142857 |
0.0010983474 |
2.701167 |
UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 |
2 |
221 |
0.007142857 |
0.0011185006 |
2.674935 |
UAUAUA |
AUAUAU,UAUAUA |
| SYNCRIP |
13 |
1041 |
0.033333333 |
0.0052498992 |
2.666604 |
AAAAAA,UUUUUU |
AAAAAA,UUUUUU |
| RBMS3 |
6 |
562 |
0.016666667 |
0.0028365578 |
2.554752 |
AAUAUA,CAUAUA,CUAUAU,UAUAUA,UAUAUC |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPU |
13 |
1136 |
0.033333333 |
0.0057285369 |
2.540727 |
AAAAAA,UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| RALY |
12 |
1117 |
0.030952381 |
0.0056328094 |
2.458124 |
UUUUUC,UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| U2AF2 |
11 |
1071 |
0.028571429 |
0.0054010480 |
2.403262 |
UUUUUC,UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 |
14 |
1381 |
0.035714286 |
0.0069629182 |
2.358737 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PABPC4 |
4 |
462 |
0.011904762 |
0.0023327287 |
2.351448 |
AAAAAA |
AAAAAA,AAAAAG |
| CPEB2 |
15 |
1487 |
0.038095238 |
0.0074969770 |
2.345230 |
AUUUUU,CAUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| PUM2 |
7 |
764 |
0.019047619 |
0.0038542926 |
2.305073 |
GUAGAU,GUAUAU,UAAAUA,UACAUC,UAGAUA,UAUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| ELAVL3 |
19 |
1928 |
0.047619048 |
0.0097188634 |
2.292679 |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| PABPC3 |
2 |
310 |
0.007142857 |
0.0015669085 |
2.188580 |
AAAAAC,AAAACA |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| KHDRBS2 |
9 |
1051 |
0.023809524 |
0.0053002821 |
2.167398 |
UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPC |
18 |
2006 |
0.045238095 |
0.0101118501 |
2.161491 |
AUUUUU,CUUUUU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| FUS |
15 |
1711 |
0.038095238 |
0.0086255542 |
2.142922 |
AAAAAA,CGGUGA,UGGUGG,UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPLL |
6 |
748 |
0.016666667 |
0.0037736800 |
2.142922 |
ACACAC,ACAUAC,CACACA,CAUACA |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| SART3 |
6 |
777 |
0.016666667 |
0.0039197904 |
2.088117 |
AAAAAA,AAAAAC,GAAAAA |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ELAVL4 |
25 |
2916 |
0.061904762 |
0.0146966949 |
2.074559 |
AAAAAA,AUUUAU,UAUUUA,UAUUUU,UCUAAU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS1 |
16 |
1945 |
0.040476190 |
0.0098045143 |
2.045555 |
AUUUAC,UAAAAA,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| A1CF |
4 |
598 |
0.011904762 |
0.0030179363 |
1.979904 |
AGUAUA,AUCAGU,CAGUAU,UCAGUA |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| IGF2BP3 |
10 |
1348 |
0.026190476 |
0.0067966546 |
1.946146 |
AAAAAC,AAAACA,AAACAC,AACACA,ACACAC,ACAUAC,CACACA,CAUACA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| KHDRBS3 |
12 |
1646 |
0.030952381 |
0.0082980653 |
1.899203 |
AAAUAA,UAAAUA,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPL |
10 |
1419 |
0.026190476 |
0.0071543732 |
1.872145 |
AAACAC,AAAUAA,AACACA,ACACAC,ACAUAC,CACACA,CAUACA |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| ZFP36L2 |
5 |
774 |
0.014285714 |
0.0039046755 |
1.871299 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| NOVA1 |
10 |
1428 |
0.026190476 |
0.0071997179 |
1.863030 |
UUCAUU,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 |
10 |
1434 |
0.026190476 |
0.0072299476 |
1.856985 |
UAGAUC,UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| YBX2 |
1 |
263 |
0.004761905 |
0.0013301088 |
1.839994 |
ACAUCU |
AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPD |
10 |
1488 |
0.026190476 |
0.0075020153 |
1.803692 |
AAAAAA,AAUUUA,AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| AGO2 |
4 |
677 |
0.011904762 |
0.0034159613 |
1.801175 |
AAAAAA |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ELAVL1 |
23 |
3309 |
0.057142857 |
0.0166767432 |
1.776736 |
AUUUAU,UAUUUA,UAUUUU,UGGUUU,UGUUUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PPIE |
64 |
9262 |
0.154761905 |
0.0466696896 |
1.729493 |
AAAAAA,AAAAAU,AAAAUU,AAAUAA,AAAUAU,AAAUUU,AAUAUA,AAUUUA,AAUUUU,AUAUAA,AUAUUU,AUUAUU,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAUA,UAAUUU,UAUAAU,UAUAUA,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| TIAL1 |
38 |
5597 |
0.092857143 |
0.0282043531 |
1.719095 |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUU,CUUUUU,GUUUUC,GUUUUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 |
34 |
5140 |
0.083333333 |
0.0259018541 |
1.685838 |
AUUUUC,AUUUUU,CUUUUU,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 |
65 |
9826 |
0.157142857 |
0.0495112858 |
1.666247 |
AAUUUA,AAUUUU,AUAUUU,AUUAUU,AUUUAC,AUUUAU,AUUUUA,AUUUUC,AUUUUU,CAUUUU,CUUUCU,CUUUUU,GUUUUC,GUUUUU,UAAUUU,UACUUU,UAUAUA,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCAUUU,UUACUU,UUAUAC,UUAUAU,UUAUUU,UUCAUU,UUCUUU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| DAZAP1 |
18 |
2878 |
0.045238095 |
0.0145052398 |
1.640964 |
AAAAAA,AAUUUA,AGGUUA,AGUAUA,UAGGUU,UAGUAA,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| RBM4 |
6 |
1094 |
0.016666667 |
0.0055169287 |
1.595028 |
CCUCUU,CCUUCC,CUUCCU,CUUCUU,UUCUUG |
CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PABPC1 |
7 |
1321 |
0.019047619 |
0.0066606207 |
1.515882 |
AAAAAA,AAAAAC,CAAAUA,GAAAAA |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| PTBP1 |
42 |
8003 |
0.102380952 |
0.0403264813 |
1.344148 |
ACUUUC,ACUUUU,AUUUUC,AUUUUU,CAUCUU,CCUCUU,CCUUCC,CUCCCC,CUCUUC,CUUCCU,CUUCUU,CUUUCU,CUUUUU,UACUUU,UAUUUU,UCCUCU,UCUCUC,UUACUU,UUAUUU,UUCUCU,UUCUUG,UUCUUU,UUUCUU,UUUUCU,UUUUUC,UUUUUU |
ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
| IGF2BP2 |
10 |
2163 |
0.026190476 |
0.0109028617 |
1.264335 |
AAAAAC,AAAACA,AAACAC,AACACA,ACACAC,ACAUAC,CACACA,CAUACA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| PUM1 |
10 |
2172 |
0.026190476 |
0.0109482064 |
1.258348 |
AAUGUU,GUAGAU,GUAUAU,UAAAUA,UAAUGU,UACAUC,UAGAUA,UAUAUA,UUAAUG |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPK |
14 |
3244 |
0.035714286 |
0.0163492543 |
1.127276 |
AAAAAA,UCCCAC,UUUUUU |
AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| QKI |
3 |
904 |
0.009523810 |
0.0045596534 |
1.062615 |
ACACAC,UCUAAU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PCBP2 |
17 |
4164 |
0.042857143 |
0.0209844821 |
1.030213 |
ACAUUA,ACCAAA,CCUCCC,CCUUCC,CUCCCC,CUUAAA,CUUCCU,UUUUUU |
AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |