circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000159140:+:21:33567156:33569087
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000159140:+:21:33567156:33569087 | ENSG00000159140 | MSTRG.21290.1 | + | 21 | 33567157 | 33569087 | 228 | CCUGUGGACAUCUCUACAGCAAUGAGUGAACGGGCACUUGCUCAGAAAAGACUCAGUGAGAAUGCAUUUGAUCUUGAAGCCAUGAGCAUGUUAAAUAGAGCUCAGGAAAGGAUUGAUGCCUGGGCUCAGCUGAACUCUAUUCCUGGCCAGUUCACAGGAAGUACAGGAGUACAGGUUUUGACACAAGAACAGUUGGCCAAUACUGGUGCCCAAGCCUGGAUUAAAAAGCCUGUGGACAUCUCUACAGCAAUGAGUGAACGGGCACUUGCUCAGAAAAG | circ |
| ENSG00000159140:+:21:33567156:33569087 | ENSG00000159140 | MSTRG.21290.1 | + | 21 | 33567157 | 33569087 | 22 | GGAUUAAAAAGCCUGUGGACAU | bsj |
| ENSG00000159140:+:21:33567156:33569087 | ENSG00000159140 | MSTRG.21290.1 | + | 21 | 33566957 | 33567166 | 210 | AUACUUUGUAAUUUAUCUUAAUCCUCUUCUGUUUUCCAAGAUUACUUUCUCUUUCUGAGUCUUCUCCCUGUUUUCCAAGAUUAUUAGUAUUAUAAGACUAUUAUUUUGUUUUUGAUAUGUAGCAACUAAAGAUAUGAGUACUUUUCAGAAUUUAGGGGACUAUGGUAAUUUGAGAUAAUUAUCUCUGUUUUCUUUCUUAGCCUGUGGACA | ie_up |
| ENSG00000159140:+:21:33567156:33569087 | ENSG00000159140 | MSTRG.21290.1 | + | 21 | 33569078 | 33569287 | 210 | GAUUAAAAAGGUACACAGUAUAUGCACAUAUGAAAGUGUCGAACAAAAAGUUUAAAUUUUUUGGAAACUGUUUAUUAAAACUGCAUGAAAAGACCAGUGACUCUAACCAAAGUUCCAAGUAAAUUUGCUAUGUGUAUAUGCUCCAGGGCGGACACUGAAGACGGACUGGAUUCAUGAGAAAAUGGCAAAUGUAUAAAUGUGGCUCAAGAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
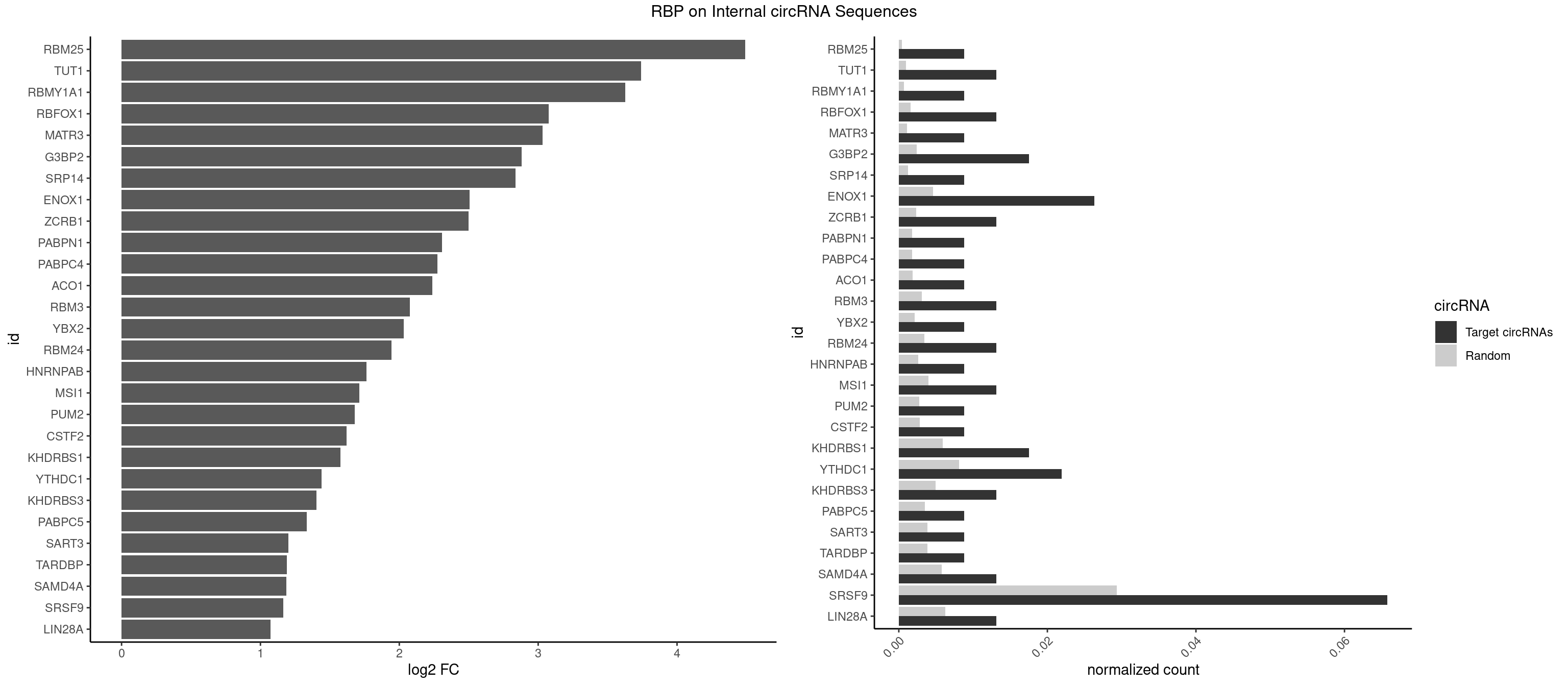
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 268 | 0.00877193 | 0.0003898359 | 4.491956 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| TUT1 | 2 | 678 | 0.01315789 | 0.0009840095 | 3.741113 | AAUACU,CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBMY1A1 | 1 | 489 | 0.00877193 | 0.0007101099 | 3.626780 | ACAAGA | ACAAGA,CAAGAC |
| RBFOX1 | 2 | 1077 | 0.01315789 | 0.0015622419 | 3.074239 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 1 | 739 | 0.00877193 | 0.0010724109 | 3.032036 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| G3BP2 | 3 | 1644 | 0.01754386 | 0.0023839405 | 2.879546 | AGGAUU,GGAUUA,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SRP14 | 1 | 847 | 0.00877193 | 0.0012289250 | 2.835497 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ENOX1 | 5 | 3195 | 0.02631579 | 0.0046316558 | 2.506329 | AGUACA,GUACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ZCRB1 | 2 | 1605 | 0.01315789 | 0.0023274215 | 2.499124 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPN1 | 1 | 1222 | 0.00877193 | 0.0017723764 | 2.307209 | AAAAGA | AAAAGA,AGAAGA |
| PABPC4 | 1 | 1251 | 0.00877193 | 0.0018144033 | 2.273399 | AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 1 | 1283 | 0.00877193 | 0.0018607779 | 2.236988 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM3 | 2 | 2152 | 0.01315789 | 0.0031201361 | 2.076248 | AAGACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| YBX2 | 1 | 1480 | 0.00877193 | 0.0021462711 | 2.031062 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBM24 | 2 | 2357 | 0.01315789 | 0.0034172229 | 1.945032 | GAGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| HNRNPAB | 1 | 1782 | 0.00877193 | 0.0025839306 | 1.763327 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MSI1 | 2 | 2770 | 0.01315789 | 0.0040157442 | 1.712189 | AGGAAG,AGUUGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| PUM2 | 1 | 1890 | 0.00877193 | 0.0027404447 | 1.678484 | UAAAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CSTF2 | 1 | 1967 | 0.00877193 | 0.0028520334 | 1.620903 | GUUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| KHDRBS1 | 3 | 4064 | 0.01754386 | 0.0058910141 | 1.574378 | UAAAAA,UUAAAA,UUAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YTHDC1 | 4 | 5576 | 0.02192982 | 0.0080822104 | 1.440072 | GAAUGC,GAGUAC,UGAUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| KHDRBS3 | 2 | 3429 | 0.01315789 | 0.0049707696 | 1.404388 | AUUAAA,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| PABPC5 | 1 | 2400 | 0.00877193 | 0.0034795387 | 1.333998 | AGAAAA | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SART3 | 1 | 2634 | 0.00877193 | 0.0038186524 | 1.199831 | AGAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TARDBP | 1 | 2654 | 0.00877193 | 0.0038476365 | 1.188922 | GUUUUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SAMD4A | 2 | 3992 | 0.01315789 | 0.0057866714 | 1.185123 | CGGGCA,CUGGCC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SRSF9 | 14 | 20254 | 0.06578947 | 0.0293536261 | 1.164318 | AGGAAA,AUGAGC,CUGGAU,GAAGCC,GAUGCC,GGAAAG,GGUGCC,UGAACG,UGAAGC,UGAGAA,UGAGCA,UGAUGC,UGGAUU,UGGUGC | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| LIN28A | 2 | 4315 | 0.01315789 | 0.0062547643 | 1.072901 | AGGAGU,GGAGUA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
RBP on BSJ (Exon Only)
Plot
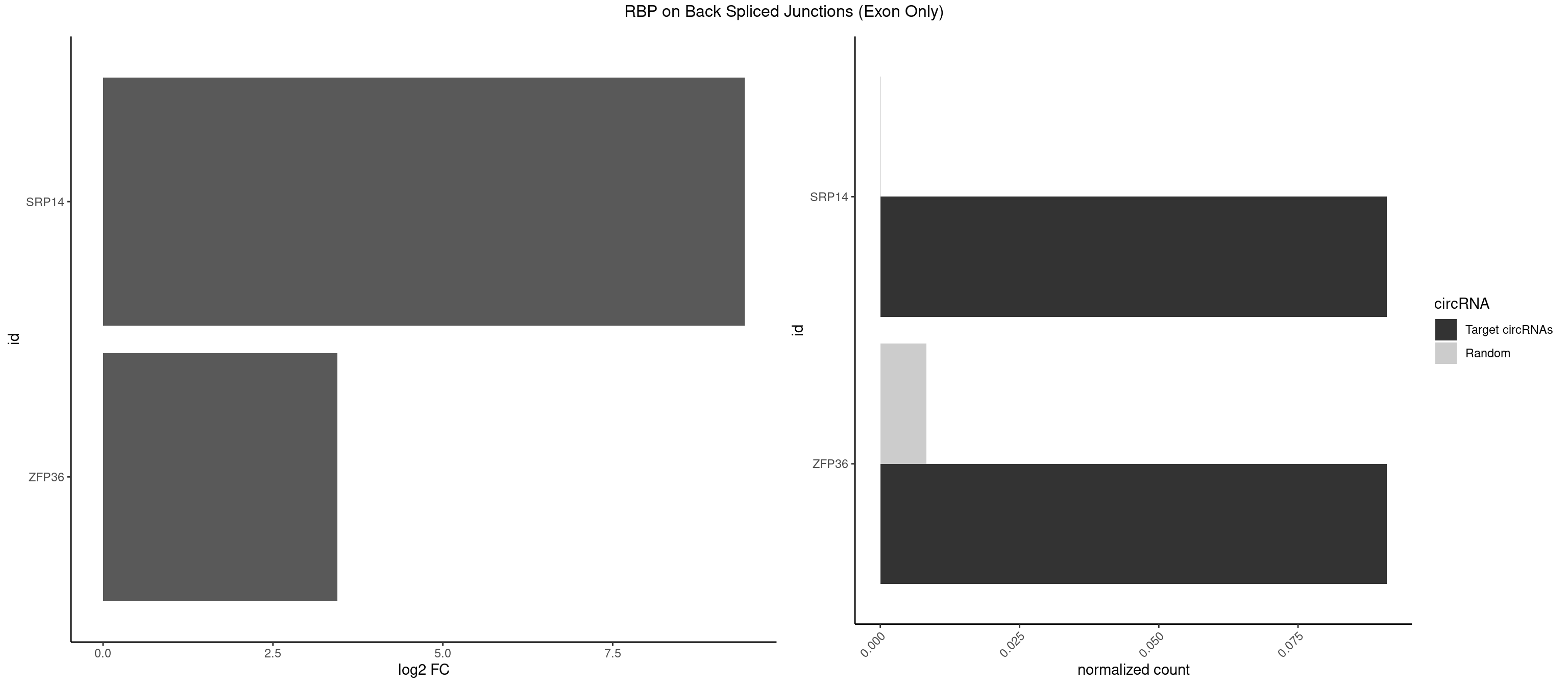
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 1 | 1 | 0.09090909 | 0.0001309929 | 9.438792 | GCCUGU | GCCUGU |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | AAAAGC | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
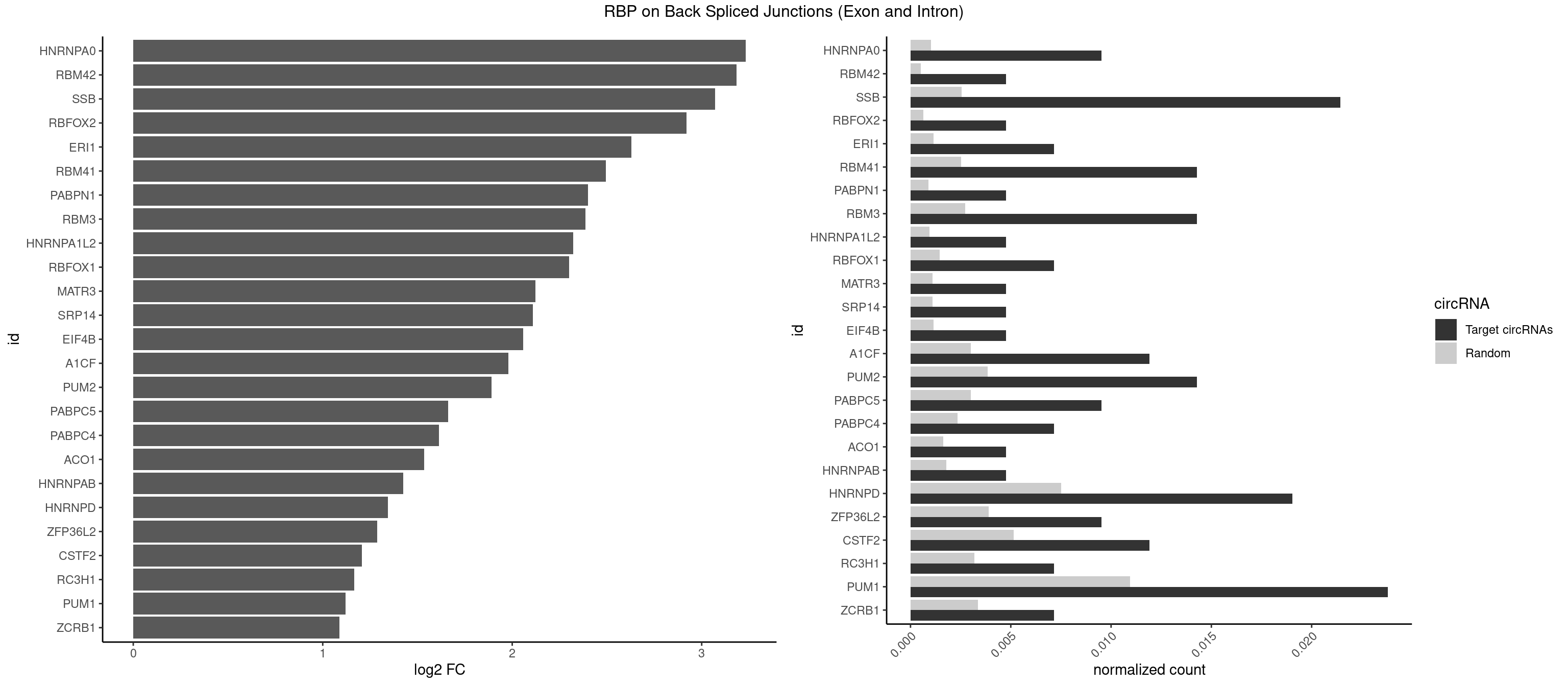
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| SSB | 8 | 505 | 0.021428571 | 0.0025493753 | 3.071320 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | AUACUU,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBM3 | 5 | 541 | 0.014285714 | 0.0027307537 | 2.387202 | AAAACU,AAGACG,AAGACU,AGACUA,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGA,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUA | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AGUAUA,AUAAUU,CAGUAU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PUM2 | 5 | 764 | 0.014285714 | 0.0038542926 | 1.890035 | GUAAAU,GUAUAU,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAA,GAAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAAGAC | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| HNRNPD | 7 | 1488 | 0.019047619 | 0.0075020153 | 1.344261 | AAUUUA,AGAUAU,AUUUAU,UUAGGG,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PUM1 | 9 | 2172 | 0.023809524 | 0.0109482064 | 1.120844 | AGAAUU,AGAUAA,CAGAAU,GUAAAU,GUAUAU,UGUAAU,UGUAUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.