circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000107829_ENSG00000224817:-:10:101667885:101673673
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000107829_ENSG00000224817:-:10:101667885:101673673 | ENSG00000107829_ENSG00000224817 | MSTRG.4529.1 | - | 10 | 101667886 | 101673673 | 414 | UCAGAUGCCCUGGAUGCAGCUAGAGGAUGAUUCUCUGUACAUAUCCCAGGCUAAUUUCAUCCUGGCCUACCAGUUCCGUCCAGAUGGUGCCAGCUUGAAUCGUCGGCCUCUGGGAGUCUUUGCUGGGCAUGAUGAGGACGUUUGCCACUUUGUGCUGGCCAACUCGCAUAUUGUUAGUGCAGGAGGGGAUGGGAAGAUUGGCAUUCAUAAGAUUCACAGCACCUUCACUGUCAAGUACUCGGCUCAUGAACAGGAGGUGAACUGUGUGGAUUGCAAAGGGGGCAUCAUUGUGAGUGGCUCCAGGGACAGGACGGCCAAGGUGUGGCCUUUGGCCUCAGGCCGGCUGGGGCAGUGCUUACACACCAUCCAGACUGAAGACCGAGUCUGGUCCAUUGCUAUCAGCCCAUUACUCAGUCAGAUGCCCUGGAUGCAGCUAGAGGAUGAUUCUCUGUACAUAUCCCAGG | circ |
| ENSG00000107829_ENSG00000224817:-:10:101667885:101673673 | ENSG00000107829_ENSG00000224817 | MSTRG.4529.1 | - | 10 | 101667886 | 101673673 | 22 | CCAUUACUCAGUCAGAUGCCCU | bsj |
| ENSG00000107829_ENSG00000224817:-:10:101667885:101673673 | ENSG00000107829_ENSG00000224817 | MSTRG.4529.1 | - | 10 | 101673664 | 101673873 | 210 | AUCACCACAAGGCUCAGUUACUCUAGUUUUCUGAGGAUAGCUGAUGGGCAGGUAAUUUUGAAGGAGGGUAUCAAGUAUAGAUAAUAAGCCUUGUUGGUUAUCCUUACUAAGUUGGGGAUUGGUCAGCUUCUUCAAAGAUUGGAGUUGAGUUUUCAUACUGUAUCUUGAUGACUUUUAAAUUUUCUUUUCCAUCAUUUCAGUCAGAUGCCC | ie_up |
| ENSG00000107829_ENSG00000224817:-:10:101667885:101673673 | ENSG00000107829_ENSG00000224817 | MSTRG.4529.1 | - | 10 | 101667686 | 101667895 | 210 | CAUUACUCAGGUAAGGGAGACAUAUUUGGAUGUGGUUUUGUCCUGUAUAAUAAUGCUUUUAUUUUACUAGAUCAGGUGAGCUAGAAAAUCUUCCUGUUGUGUAUUCUAGGUUGCUUUGAGUCACUGUCUUGGAGAUUUUUUAAUCCUCCAGAGAUUUGAGUUAUUUAGGCCACUUCAUAUAUGCGUGGGAUUGAGAUAACCUUCUAAAUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
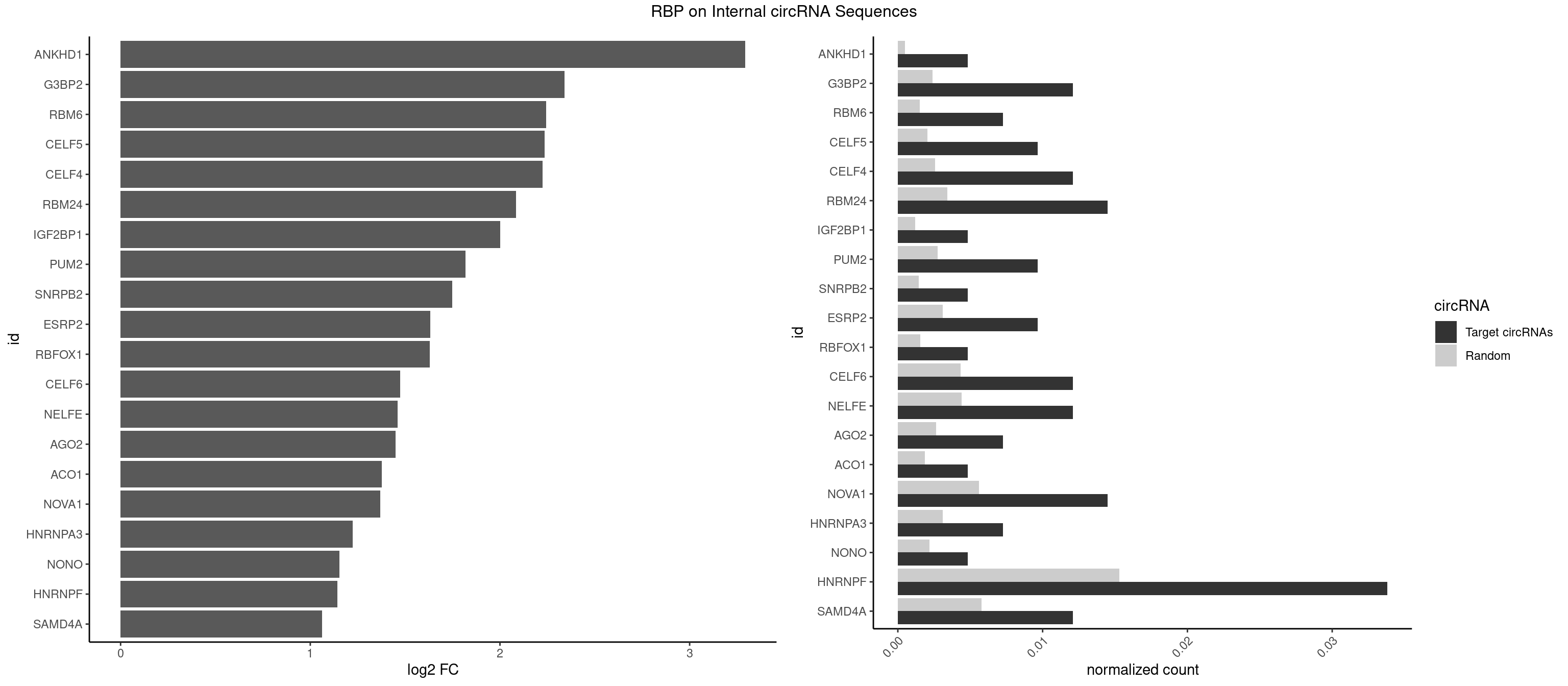
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 339 | 0.004830918 | 0.0004927293 | 3.293430 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| G3BP2 | 4 | 1644 | 0.012077295 | 0.0023839405 | 2.340877 | AGGAUG,GGAUGA,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM6 | 2 | 1054 | 0.007246377 | 0.0015289102 | 2.244756 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CELF5 | 3 | 1415 | 0.009661836 | 0.0020520728 | 2.235215 | GUGUGG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 1782 | 0.012077295 | 0.0025839306 | 2.224658 | GGUGUG,GUGUGG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM24 | 5 | 2357 | 0.014492754 | 0.0034172229 | 2.084435 | GAGUGG,GUGUGG,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| IGF2BP1 | 1 | 831 | 0.004830918 | 0.0012057377 | 2.002381 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PUM2 | 3 | 1890 | 0.009661836 | 0.0027404447 | 1.817887 | GUACAU,UACAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| SNRPB2 | 1 | 991 | 0.004830918 | 0.0014376103 | 1.748625 | AUUGCA | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ESRP2 | 3 | 2150 | 0.009661836 | 0.0031172377 | 1.632029 | GGGAAG,GGGGAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBFOX1 | 1 | 1077 | 0.004830918 | 0.0015622419 | 1.628680 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| CELF6 | 4 | 2997 | 0.012077295 | 0.0043447134 | 1.474964 | GUGUGG,UGUGAG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| NELFE | 4 | 3028 | 0.012077295 | 0.0043896388 | 1.460123 | CUCUGG,GGCUAA,UCUCUG,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| AGO2 | 2 | 1830 | 0.007246377 | 0.0026534924 | 1.449367 | AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ACO1 | 1 | 1283 | 0.004830918 | 0.0018607779 | 1.376391 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| NOVA1 | 5 | 3872 | 0.014492754 | 0.0056127669 | 1.368548 | AGCACC,AUUCAU,CAGUCA,CAUUCA,UCAGUC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPA3 | 2 | 2140 | 0.007246377 | 0.0031027457 | 1.223714 | CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| NONO | 1 | 1498 | 0.004830918 | 0.0021723567 | 1.153036 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| HNRNPF | 13 | 10561 | 0.033816425 | 0.0153064921 | 1.143580 | AAGGGG,AAGGUG,AGGGGA,AUGGGA,CUGGGG,GAGGGG,GAUGGG,GGAGGG,GGAUGG,GGGAAG,GGGAUG,GGGGGC,UGGGAA | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| SAMD4A | 4 | 3992 | 0.012077295 | 0.0057866714 | 1.061492 | CUGGCC,CUGGUC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
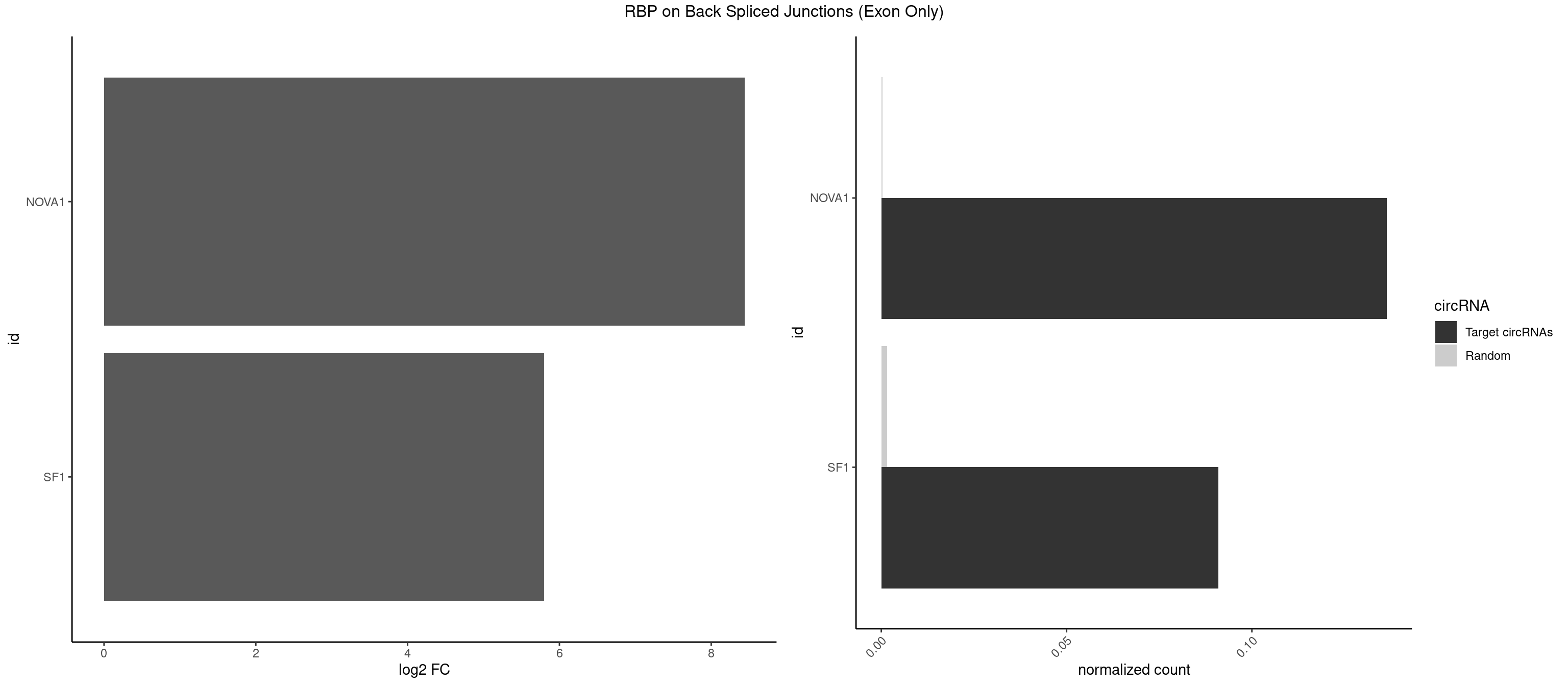
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NOVA1 | 2 | 5 | 0.13636364 | 0.0003929788 | 8.438792 | CAGUCA,UCAGUC | ACCACC,CAGUCA,CAUUCA,UCAGUC |
| SF1 | 1 | 24 | 0.09090909 | 0.0016374116 | 5.794936 | CAGUCA | ACAGAC,ACAGUC,AGUAAG,AUACUA,CACAGA,CACUGA,CAGUCA,GCUGAC,GCUGCC,UACUGA,UGCUAA,UGCUGA,UGCUGC |
RBP on BSJ (Exon and Intron)
Plot
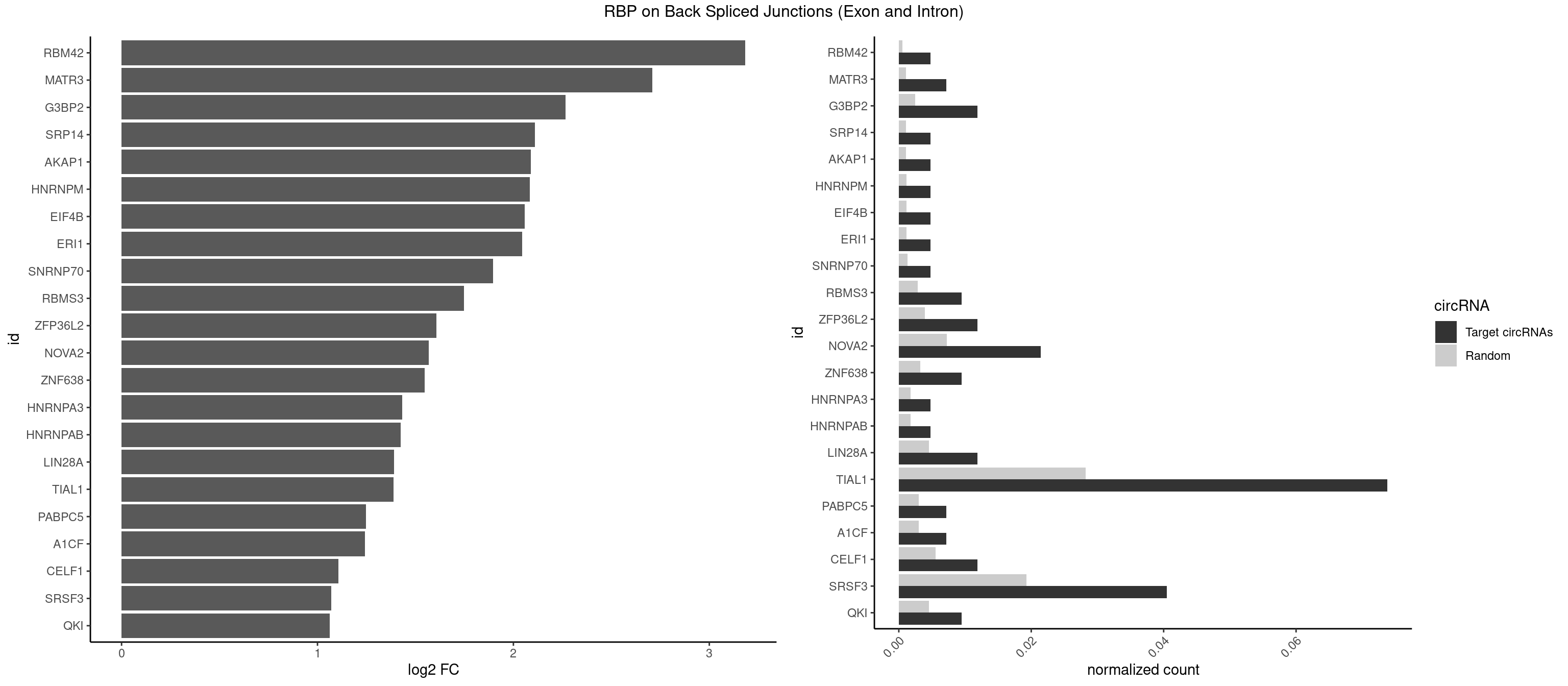
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUA,GGAUAG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | CUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CAUAUA,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| NOVA2 | 8 | 1434 | 0.021428571 | 0.0072299476 | 1.567479 | AGACAU,AGAUCA,AUCACC,CUAGAU,GAGACA,GAGUCA,UAGAUC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GUUGGU,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGAU,GGAGGG,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| TIAL1 | 30 | 5597 | 0.073809524 | 0.0282043531 | 1.387889 | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CUUUUA,CUUUUC,GUUUUC,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AGUAUA,GAUCAG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | CUGUCU,GUUGUG,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SRSF3 | 16 | 3825 | 0.040476190 | 0.0192765014 | 1.070230 | ACUUCA,AGAGAU,AUCUUC,CACCAC,CACUUC,CAGAGA,CAUCAU,CUUCAA,CUUCAU,CUUCUU,GAGAUU,UAUCAA,UCUUCA,UCUUCC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | UAACCU,UACUCA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.