circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000288622:+:5:314416:428006
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000288622:+:5:314416:428006 | ENSG00000288622 | MSTRG.25844.34 | + | 5 | 314417 | 428006 | 1098 | AGGUUGACGGAUAUAUUCAGACGUUACGACACGGAUCAGGACGGCUGGAUUCAGGUGUCGUACGAACAGUACCUGUCCAUGGUCUUCAGUAUCGUAUGACCCUGGCCUCUCGUGAAGAGCAGCACAACAUGGAAAGAGCCAAAAUGUCACAGUUCCUAUCUGUGAGGGAAUGGAGCACAGGCCGAGGACGAUGAUCCCGCCGGGGGAGUGCACGUACGCGGGCCGGAAGCGGAGGAGGCCCCUGCAGAAACAGAGGCCCGCCGUGGGGGCAGAGAAGUCCAACCCCUCCAAGCGACACCGGGACCGCCUCAACGCCGAGUUGGACCACCUGGCCAGCCUGCUGCCGUUCCCGCCUGACAUCAUCUCCAAGCUGGACAAGCUUUCUGUCCUGCGCCUCAGUGUCAGUUACCUCCGGGUGAAGAGCUUCUUCCAAGUCGUGCAGGAGCAGAGCUCACGGCAGCCUGCGGCCGGCGCCCCCUCGCCCGGAGACAGCUGUCCUCUUGCAGGGUCUGCCGUGCUGGAGGGAAGGCUGCUGUUGGAGUCUCUUAAUGGCUUUGCUCUGGUCGUGAGUGCAGAAGGGACGAUAUUUUAUGCAUCAGCAACGAUCGUGGACUAUCUGGGCUUCCAUCAGACGGAUGUAAUGCACCAGAACAUUUAUGACUACAUCCACGUGGACGACCGCCAGGACUUCUGCCGGCAGCUCCACUGGGCCAUGGACCCUCCCCAGGUGGUGUUUGGGCAGCCCCCGCCCUUGGAGACAGGAGAUGAUGCUAUCCUGGGGAGGCUGCUCAGGGCCCAGGAGUGGGGCACAGGCACGCCCACCGAGUACUCGGCCUUCCUGACCCGCUGCUUCAUCUGCCGUGUGCGCUGCCUGCUGGACAGCACCUCGGGCUUCCUGACGAUGCAGUUUCAAGGAAAACUAAAAUUCCUGUUUGGACAGAAGAAGAAGGCGCCGUCAGGAGCCAUGCUCCCGCCGCGGCUGUCGCUGUUCUGCAUUGCGGCACCCGUUCUCCUCCCCUCCGCAGCGGAGAUGAAAAUGAGGAGCGCGCUCCUGAGGGCAAAACCCAGAGCAGACACCGCAGCCACCGCGGAUGCAAAAGGUUGACGGAUAUAUUCAGACGUUACGACACGGAUCAGGACGGCUGGAU | circ |
| ENSG00000288622:+:5:314416:428006 | ENSG00000288622 | MSTRG.25844.34 | + | 5 | 314417 | 428006 | 22 | GCGGAUGCAAAAGGUUGACGGA | bsj |
| ENSG00000288622:+:5:314416:428006 | ENSG00000288622 | MSTRG.25844.34 | + | 5 | 314217 | 314426 | 210 | AGCCCUGUGUUCUUCGUUCUUCGCACUCCCACCGUCCGUGUGAACAGCUCCAGCCCCACCUGCGCCUCCCUGUGCUGGGCUCCAUCAGGGAGCCCAGAAGACGUGUGUGCUUCUGAAAUUGGGUCCCUACAUGCCUUUGUCCCAGUGCACCUUGCUCCUUCCAUUUACUAUCGAGAUUUAAAUGCCUGUUUUCUCCCCAGAGGUUGACGG | ie_up |
| ENSG00000288622:+:5:314416:428006 | ENSG00000288622 | MSTRG.25844.34 | + | 5 | 427997 | 428206 | 210 | CGGAUGCAAAGUGAGUAAGACUCGCCCUUCACAGCCACAUGGUGCCUGCUGGCGGCCCCCACAAAACACCUCCACACUCAGUGUUUUCUGUGUCUUCUUUCUUCAUUUCCAGCCAGUAAUAAGUCAGUUUCGUGUCGUAAAAGUCAUUACACAACAUAGGCAGCAGCGAUUAGAUGAAACAAUGUUUUUCAAACUCGUAUUUGCAAACUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
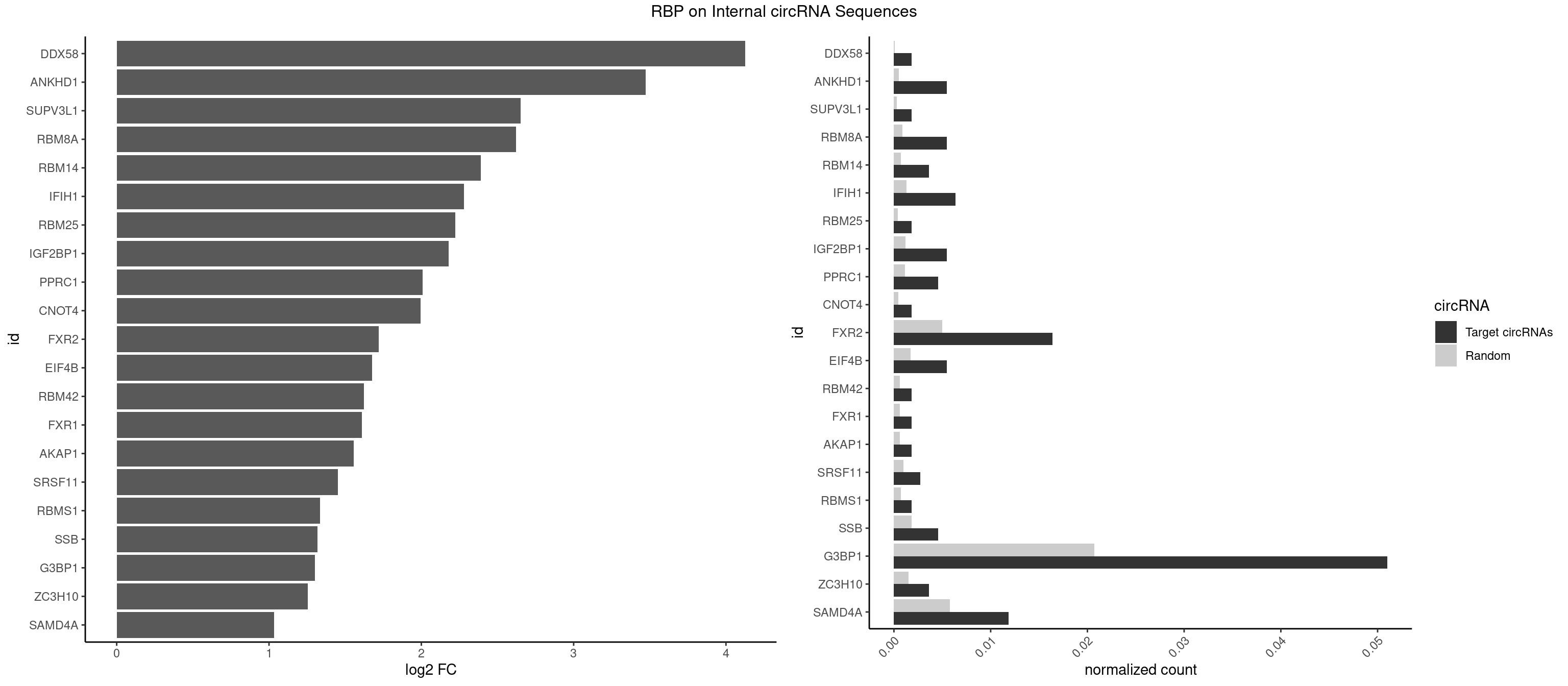
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.001821494 | 0.0001043427 | 4.125721 | GCGCGC | GCGCGC |
| ANKHD1 | 5 | 339 | 0.005464481 | 0.0004927293 | 3.471217 | AGACGU,GACGAU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SUPV3L1 | 1 | 199 | 0.001821494 | 0.0002898408 | 2.651789 | CCGCCC | CCGCCC |
| RBM8A | 5 | 611 | 0.005464481 | 0.0008869128 | 2.623220 | CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM14 | 3 | 478 | 0.003642987 | 0.0006941687 | 2.391764 | CGCGGC,CGCGGG,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| IFIH1 | 6 | 904 | 0.006375228 | 0.0013115296 | 2.281227 | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM25 | 1 | 268 | 0.001821494 | 0.0003898359 | 2.224183 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| IGF2BP1 | 5 | 831 | 0.005464481 | 0.0012057377 | 2.180168 | ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PPRC1 | 4 | 780 | 0.004553734 | 0.0011318283 | 2.008395 | CGGCGC,GCGCGC,GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| CNOT4 | 1 | 314 | 0.001821494 | 0.0004564992 | 1.996438 | GACAGA | GACAGA |
| FXR2 | 17 | 3434 | 0.016393443 | 0.0049780156 | 1.719476 | AGACAG,AGACGG,GACAAG,GACAGA,GACAGG,GACGGA,GGACAA,GGACAG,GGACGA,GGACGG,UGACGA,UGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| EIF4B | 5 | 1179 | 0.005464481 | 0.0017100607 | 1.676037 | CUUGGA,GUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM42 | 1 | 407 | 0.001821494 | 0.0005912752 | 1.623220 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.001821494 | 0.0005970720 | 1.609145 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| AKAP1 | 1 | 426 | 0.001821494 | 0.0006188101 | 1.557553 | AUAUAU | AUAUAU,UAUAUA |
| SRSF11 | 2 | 688 | 0.002732240 | 0.0009985015 | 1.452248 | AAGAAG | AAGAAG |
| RBMS1 | 1 | 497 | 0.001821494 | 0.0007217036 | 1.335644 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SSB | 4 | 1260 | 0.004553734 | 0.0018274462 | 1.317221 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| G3BP1 | 55 | 14282 | 0.051001821 | 0.0206989801 | 1.300989 | ACAGGC,ACCCCU,ACCCGC,ACGCCC,ACGCCG,AGGCAC,AGGCCC,AGGCCG,CACAGG,CACCCG,CACCGG,CACGCC,CAGGCA,CAGGCC,CCACCG,CCCACC,CCCAGG,CCCCAG,CCCCCG,CCCCGC,CCCCUC,CCCGCC,CCCUCC,CCCUCG,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCG,CUCCGC,CUCGGC,UCCGCA,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| ZC3H10 | 3 | 1053 | 0.003642987 | 0.0015274610 | 1.253986 | GAGCGC,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SAMD4A | 12 | 3992 | 0.011839709 | 0.0057866714 | 1.032828 | CGGGAC,CGGGCC,CUGGAC,CUGGCC,CUGGUC,GCGGGC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
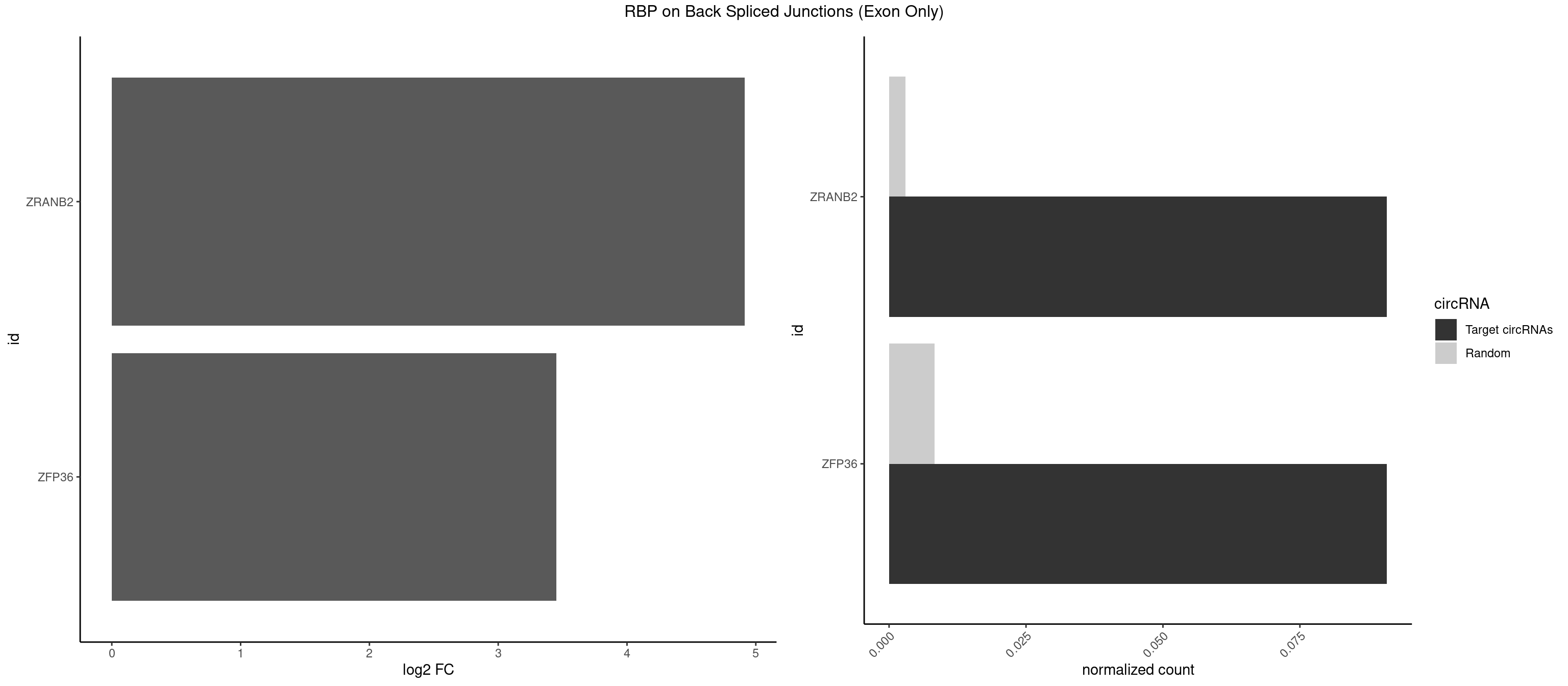
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AAAGGU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AAAAGG | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
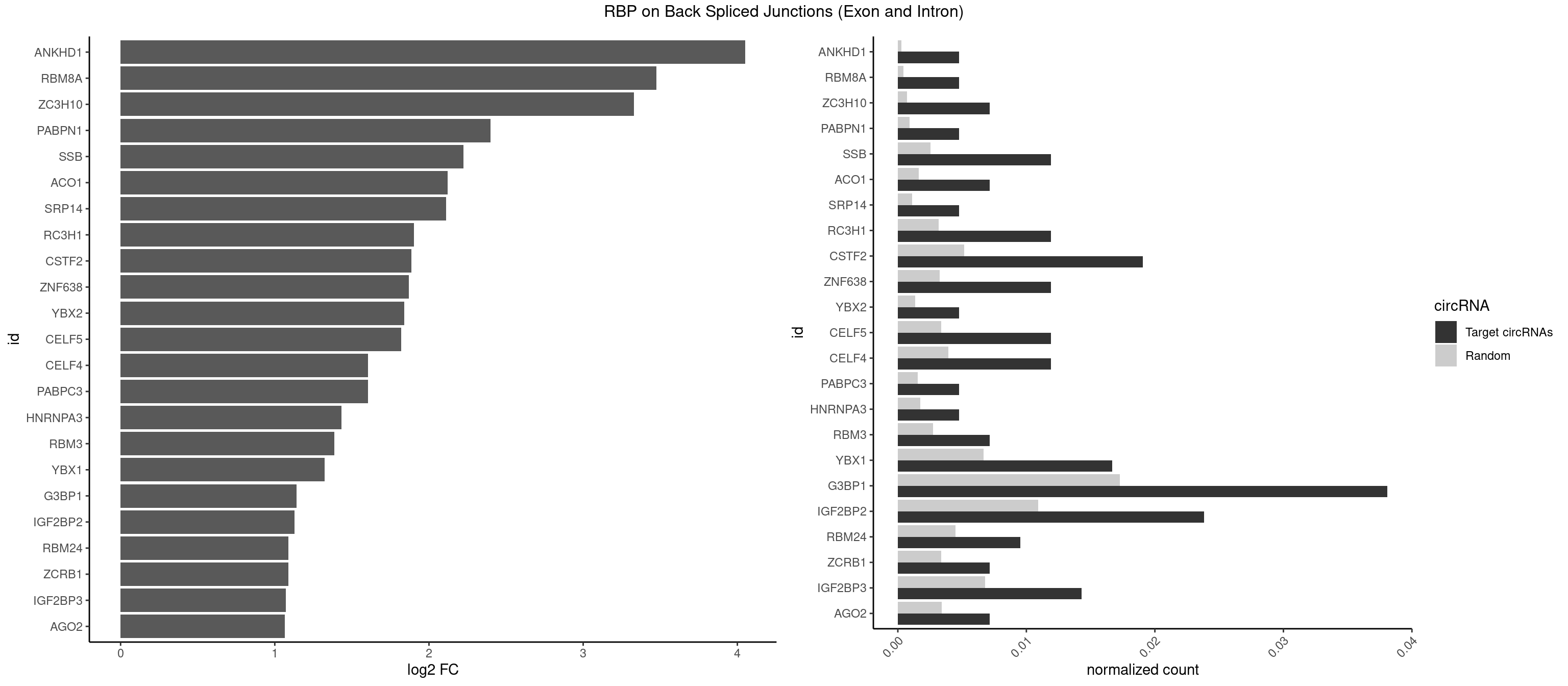
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 1 | 56 | 0.004761905 | 0.0002871826 | 4.051499 | AGACGU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| ZC3H10 | 2 | 140 | 0.007142857 | 0.0007103990 | 3.329800 | CAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AGAAGA | AAAAGA,AGAAGA |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CCCUUC,CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | CGUUCU,GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAACA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACG,AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | CCACAA,CCACAC,CCUGCG,GCCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| G3BP1 | 15 | 3431 | 0.038095238 | 0.0172914148 | 1.139555 | AGGCAG,AUAGGC,CAUAGG,CCACAC,CCACCG,CCCACA,CCCACC,CCCCAC,CCCCAG,CCCCCA,CCCUAC,CGGCCC,UAGGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| IGF2BP2 | 9 | 2163 | 0.023809524 | 0.0109028617 | 1.126832 | AAAACA,AAACAC,AAACUC,ACACUC,CAAAAC,CACUCA,CCACAC,GCAAAC,GCACUC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | GUGUGA,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAACA,AAACAC,AAACUC,ACACUC,CACUCA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAGUG,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.